 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AHYPO_001951-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Amaranthaceae; Amaranthus
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 330aa MW: 35225.4 Da PI: 8.4568 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 207.5 | 3.6e-64 | 2 | 147 | 2 | 149 |
DUF822 2 gsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeaeaagssasaspesslq 95
+++r+ptw+ErEnnkrRERrRRaiaakiy+GLR++Gnyklpk++DnneVlkAL+reAGw+ve+DGttyrkg+kp e ++++g sas+sp+ss+
AHYPO_001951-RA 2 TGTRMPTWRERENNKRRERRRRAIAAKIYSGLRMYGNYKLPKHCDNNEVLKALAREAGWIVEEDGTTYRKGCKPVEPMDIIGGSASVSPCSSYP 95
689******************************************************************************************* PP
DUF822 96 sslkssalaspvesysaspksssfpspssldsislasa....asllpvlsvlslvsss 149
+ +sy++sp+sssfpsp+++ ++++ + +sl+p+l++ls++ ss
AHYPO_001951-RA 96 ------VSPRAGASYNPSPASSSFPSPARSYCVPNVNGttdpNSLIPWLKNLSSSGSS 147
......44566789****************999887667788*********9985443 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 9.2E-64 | 3 | 142 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 330 aa Download sequence Send to blast |
MTGTRMPTWR ERENNKRRER RRRAIAAKIY SGLRMYGNYK LPKHCDNNEV LKALAREAGW 60 IVEEDGTTYR KGCKPVEPMD IIGGSASVSP CSSYPVSPRA GASYNPSPAS SSFPSPARSY 120 CVPNVNGTTD PNSLIPWLKN LSSSGSSSTS SKLPYIYIPG GSISAPVTPP LSSPTCRTPR 180 TKNDWDDSNV VPPWANPANY SLPSSTPQSP GRQPLPEPGW LANIQIPQGA PTSPTFSLVS 240 SNPFASKDEV MAGSGSRMWT PGQSGTCSPI VATGSDRTAD VPMSDGIAAE FAFGNGMTGG 300 LVKAWEGEII HEECVPDDLE LTLGSSRTR* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5zd4_A | 7e-24 | 5 | 86 | 372 | 452 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_B | 7e-24 | 5 | 86 | 372 | 452 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_C | 7e-24 | 5 | 86 | 372 | 452 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_D | 7e-24 | 5 | 86 | 372 | 452 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00248 | DAP | Transfer from AT1G78700 | Download |
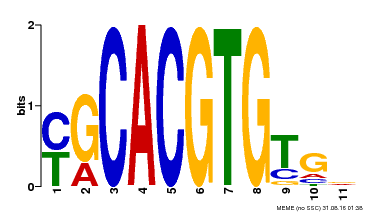
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010665681.1 | 0.0 | PREDICTED: BES1/BZR1 homolog protein 4 | ||||
| Swissprot | Q9ZV88 | 1e-114 | BEH4_ARATH; BES1/BZR1 homolog protein 4 | ||||
| TrEMBL | A0A0J8B9H3 | 0.0 | A0A0J8B9H3_BETVU; Uncharacterized protein | ||||
| STRING | XP_010665681.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G78700.1 | 6e-96 | BES1/BZR1 homolog 4 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AHYPO_001951-RA |



