 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AHYPO_002079-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Amaranthaceae; Amaranthus
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 571aa MW: 62605.7 Da PI: 7.3234 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 97 | 1.2e-30 | 305 | 363 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-ST.T---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsa.gCpvkkkversaedpkvveitYegeHnhe 59
+Dg++WrKYGqK+ kg+++pr+YYrCt+a gCpv+k+v+r+aed +++++tYeg+Hnh+
AHYPO_002079-RA 305 SDGCQWRKYGQKMAKGNPCPRAYYRCTMAvGCPVRKQVQRCAEDRTILITTYEGTHNHP 363
7*********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 4.0E-33 | 289 | 365 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 7.32E-28 | 297 | 365 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 29.562 | 299 | 365 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.8E-36 | 304 | 364 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.3E-26 | 306 | 363 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 571 aa Download sequence Send to blast |
MDGGGWGLAL ESSSSSSDHN KLITPNNFSI HHNINKNEMF KGLEFPINLK RRSGNQMENH 60 DHHNRDDHNR DDRNGKTVVD VLNFFPESKE ITGLHLHTNN TGSDQSTIDD SASSDIEDKK 120 NKNELTILQG ELQRMNTENE RLKILLQQVH TNYMHLENQM KQANFEIAKN KACTHRKVDE 180 MQTMQGQFLN LAPKDNNEVD ELTSKTVSDD EKTIERSSSP QNTIEVVSVR TNNKDVMLSN 240 YDHGKRIREQ SVEELEAKRW QNKVPQLMNN VLKPTSPPID HSANEATMRK VRVSVRARSE 300 APMISDGCQW RKYGQKMAKG NPCPRAYYRC TMAVGCPVRK QVQRCAEDRT ILITTYEGTH 360 NHPLPPAAVP MVETTSSAAT MLLSGSMPSG DGLMNPNFLA ARTILPCSSS VATISASAPF 420 PTVTLDLTYS PNPLQQRPSI PFPPPNAQVG PNLLGSIPQI FGQALLNSSN NQSKFSGLQT 480 STPQSLNAAH LQPSNQNAAF NETLSAATAA LTSDPNFTAA LVAALSSIIG GATRHPTENN 540 HTSNNAGGNS PSGATNNAKN DASNNNMQQQ * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2ayd_A | 5e-22 | 292 | 367 | 2 | 76 | WRKY transcription factor 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
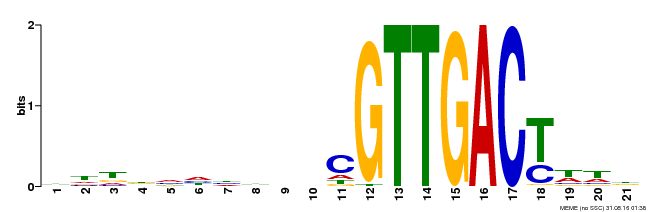
| UniProt | Transcription factor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (By similarity). Modulates phosphate homeostasis and Pi translocation by regulating PHO1 expression (PubMed:25733771). {ECO:0000250, ECO:0000269|PubMed:25733771}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00428 | DAP | Transfer from AT4G04450 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010686807.1 | 0.0 | PREDICTED: probable WRKY transcription factor 31 isoform X2 | ||||
| Swissprot | Q9XEC3 | 1e-123 | WRK42_ARATH; WRKY transcription factor 42 | ||||
| TrEMBL | A0A0K9S1P8 | 0.0 | A0A0K9S1P8_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010686806.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G04450.1 | 1e-117 | WRKY family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AHYPO_002079-RA |



