 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AHYPO_004690-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Amaranthaceae; Amaranthus
|
||||||||
| Family | TCP | ||||||||
| Protein Properties | Length: 282aa MW: 30304.4 Da PI: 8.2718 | ||||||||
| Description | TCP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | TCP | 127.7 | 1.2e-39 | 35 | 121 | 2 | 88 |
TCP 2 agkkdrhskihTkvggRdRRvRlsaecaarfFdLqdeLGfdkdsktieWLlqqakpaikeltgtssssaseceaesssssasnsssg 88
a+k+++++++hTkv+gR RR+R++a+caar+F+L++eLG+++d++ti+WLlqqa+pai+++tgt++++as+ +a++s +++ ss +
AHYPO_004690-RA 35 APKRSSNKDRHTKVDGRSRRIRMPALCAARVFQLTRELGHKTDGETIQWLLQQAEPAIIAATGTGTIPASFLTASASGATQQGSSVS 121
789******************************************************************888554444444433333 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03634 | 1.4E-34 | 40 | 276 | IPR005333 | Transcription factor, TCP |
| PROSITE profile | PS51369 | 27.714 | 41 | 95 | IPR017887 | Transcription factor TCP subgroup |
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 282 aa Download sequence Send to blast |
MDDNRNPEEK QAVTINTQSP NKEDLQKKQQ NQQLAPKRSS NKDRHTKVDG RSRRIRMPAL 60 CAARVFQLTR ELGHKTDGET IQWLLQQAEP AIIAATGTGT IPASFLTASA SGATQQGSSV 120 SLGLHSKINE LRAGIEVGNR PNWANFGDNL GRSQVGASIW PSIGSGYAQV FDNCSGTNSN 180 LVSQSSSYVT NFQLQGVGLP TGNMGLLSFT PILTRGTQIP GLQLGLSQDG VIGQLYQQQQ 240 QQQQGHEHTD SGAAAAAVSA ETVHQQQQHL QSHSNDNSRE Q* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds to the site II motif (3'-TGGGCC/T-5') in the promoter of PCNA-2 and to 3'-GCCCG/A-5' elements in the promoters of cyclin CYCB1-1 and ribosomal protein genes. {ECO:0000269|PubMed:12631321, ECO:0000269|PubMed:16123132}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00636 | PBM | Transfer from Glyma.19G095300 | Download |
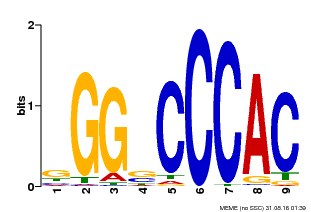
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021758425.1 | 1e-117 | transcription factor TCP20-like | ||||
| Swissprot | Q9LSD5 | 5e-56 | TCP20_ARATH; Transcription factor TCP20 | ||||
| TrEMBL | A0A0J8BC90 | 1e-115 | A0A0J8BC90_BETVU; Uncharacterized protein | ||||
| STRING | XP_006484165.1 | 4e-82 | (Citrus sinensis) | ||||
| STRING | XP_006437974.1 | 4e-82 | (Citrus clementina) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G27010.1 | 2e-54 | TCP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AHYPO_004690-RA |



