 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AHYPO_006644-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Amaranthaceae; Amaranthus
|
||||||||
| Family | ARR-B | ||||||||
| Protein Properties | Length: 622aa MW: 70202.3 Da PI: 5.2444 | ||||||||
| Description | ARR-B family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 76 | 5e-24 | 191 | 247 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLG..GsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
k+r++Wt eLH++Fveav+qLG k Pk+il++m+v+ Lt+e+v+SHLQkYRl
AHYPO_006644-RA 191 KARVVWTVELHQKFVEAVNQLGidSDSKIGPKKILDMMNVPWLTRENVASHLQKYRL 247
68*******************633345789**************************8 PP
| |||||||
| 2 | Response_reg | 76.7 | 8.2e-26 | 21 | 129 | 1 | 109 |
EEEESSSHHHHHHHHHHHHHTTCEEEEEESSHHHHHHHHHHHH..ESEEEEESSCTTSEHHHHHHHHHHHTTTSEEEEEESTTTHHHHHHHHHT CS
Response_reg 1 vlivdDeplvrellrqalekegyeevaeaddgeealellkekd..pDlillDiempgmdGlellkeireeepklpiivvtahgeeedalealka 92
vl+vdD+ + +++l+++l+k y ev+++ ++eal+ll++++ +D+++ D++mp+mdG++ll++ e +lp+i+++ ge + + + ++
AHYPO_006644-RA 21 VLVVDDDLTWLKILEKMLKKCSY-EVTTCGLASEALKLLRDRKdgFDIVISDVNMPDMDGFKLLEHVGLEM-DLPVIMMSVDGETSRVMKGVQH 112
89*********************.***************888889**********************6644.8********************* PP
TESEEEESS--HHHHHH CS
Response_reg 93 GakdflsKpfdpeelvk 109
Ga d+l Kp+ ++el +
AHYPO_006644-RA 113 GACDYLLKPIRMKELKN 129
*************9986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF52172 | 4.27E-35 | 17 | 141 | IPR011006 | CheY-like superfamily |
| Gene3D | G3DSA:3.40.50.2300 | 5.9E-44 | 18 | 173 | No hit | No description |
| SMART | SM00448 | 9.1E-31 | 19 | 131 | IPR001789 | Signal transduction response regulator, receiver domain |
| PROSITE profile | PS50110 | 42.204 | 20 | 135 | IPR001789 | Signal transduction response regulator, receiver domain |
| Pfam | PF00072 | 7.1E-23 | 21 | 129 | IPR001789 | Signal transduction response regulator, receiver domain |
| CDD | cd00156 | 4.26E-29 | 22 | 135 | No hit | No description |
| SuperFamily | SSF46689 | 2.42E-17 | 188 | 251 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 3.2E-27 | 189 | 252 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.6E-20 | 191 | 247 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 3.2E-8 | 193 | 246 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000160 | Biological Process | phosphorelay signal transduction system | ||||
| GO:0009735 | Biological Process | response to cytokinin | ||||
| GO:0010082 | Biological Process | regulation of root meristem growth | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 622 aa Download sequence Send to blast |
MVMENGFSSP RSDIFPSGLR VLVVDDDLTW LKILEKMLKK CSYEVTTCGL ASEALKLLRD 60 RKDGFDIVIS DVNMPDMDGF KLLEHVGLEM DLPVIMMSVD GETSRVMKGV QHGACDYLLK 120 PIRMKELKNI WQHVMRKRMQ EVRDIDCFED RNGTDHYDGH CGGDQMSLRK RKDVENKHDE 180 KDSSDSSTTK KARVVWTVEL HQKFVEAVNQ LGIDSDSKIG PKKILDMMNV PWLTRENVAS 240 HLQKYRLYLS RIQKGDKLEN SCGGKKNPDH SYKESSGILG LPNPSFGQVN DISSKYVYRG 300 NQLPIKTVDS IPLDSNPESS KFLPLTEHRV CFSSESSDPQ NARLGLAHTF GSLDMSIKHT 360 SFDSHVATRY PWSGEATDLQ FEPDFKALPT LDTCFNQPVE RILQQQISPD LLDPAPHVSP 420 NLLESAPHIS NDVLEPVLHV KAGRCDTGKV SYLESAPESC SVSNVDAFGR VFDTKNHNFS 480 QRVDDMKETQ PLFERFPTQF EENHLIDTQG FEPSSVGALE MKRQTSVPGS LTAETTEYEL 540 RNQILRCESP STLLGEDLIH RLIQYGDVTS KDFGFDCMGF TGCNFSENDF YNPLPLHRPK 600 LGTEYPIDWA EYPLDQGLLF S* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 3e-17 | 187 | 252 | 1 | 63 | ARR10-B |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that binds specific DNA sequence. Functions as a response regulator involved in His-to-Asp phosphorelay signal transduction system. Phosphorylation of the Asp residue in the receiver domain activates the ability of the protein to promote the transcription of target genes. May directly activate some type-A response regulators in response to cytokinins. {ECO:0000250|UniProtKB:Q940D0}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00010 | PBM | Transfer from AT1G67710 | Download |
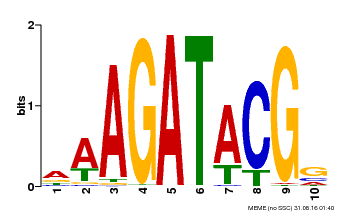
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010682390.1 | 0.0 | PREDICTED: two-component response regulator ARR11 isoform X1 | ||||
| Swissprot | Q5N6V8 | 1e-125 | ORR26_ORYSJ; Two-component response regulator ORR26 | ||||
| TrEMBL | A0A0K9QRY8 | 0.0 | A0A0K9QRY8_SPIOL; Two-component response regulator | ||||
| STRING | XP_010682390.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G67710.1 | 1e-123 | response regulator 11 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AHYPO_006644-RA |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



