 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AHYPO_008931-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Amaranthaceae; Amaranthus
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 278aa MW: 30663.4 Da PI: 6.6749 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 52.9 | 8.4e-17 | 14 | 61 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rgrWT eEd++l++++ + G g+W++ ++ g+ R++k+c++rw +yl
AHYPO_008931-RA 14 RGRWTDEEDQKLIKYILENGEGSWRSLPKNAGLLRCGKSCRLRWINYL 61
8*******************************99************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 1.6E-21 | 5 | 64 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 22.515 | 9 | 65 | IPR017930 | Myb domain |
| SMART | SM00717 | 3.3E-12 | 13 | 63 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.8E-15 | 14 | 61 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 1.71E-20 | 15 | 88 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 3.40E-10 | 16 | 61 | No hit | No description |
| PROSITE profile | PS50090 | 3.95 | 62 | 130 | IPR017877 | Myb-like domain |
| Gene3D | G3DSA:1.10.10.60 | 1.1E-7 | 65 | 88 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 18 | 66 | 137 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 278 aa Download sequence Send to blast |
MGRAPCCEKV GLKRGRWTDE EDQKLIKYIL ENGEGSWRSL PKNAGLLRCG KSCRLRWINY 60 LRSDLKRGNI TPEEEEIIIK LHTTLGNRKI HTFRRPNGEG SEEKIIINLS NVSAPPKRRG 120 GRTSRAAMQR NRLNKVMNNN NNNINIALMG PNQVGPSVPK NSTTSNGNLI FDHESELLGI 180 EDIMGLDGLL STTNEDSVME SDAGVGPNES KGPMKMGSIA EDPIHQNITC SGSSGGGSST 240 TTSSSNFEHN FDWDLDIGTV DVVGPEFKLW DEKDQIIS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 9e-12 | 12 | 88 | 5 | 80 | B-MYB |
| 1h8a_C | 2e-11 | 12 | 88 | 25 | 100 | MYB TRANSFORMING PROTEIN |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00668 | SELEX | Transfer from GRMZM2G016020 | Download |
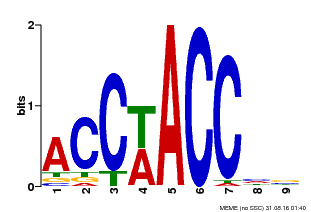
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KJ707238 | 1e-51 | KJ707238.1 Beta vulgaris subsp. vulgaris cultivar KWS2320 R2R3-MYB flavonol regulator (MYB12) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021836184.1 | 1e-108 | transcription factor MYB11 | ||||
| TrEMBL | A0A096ZXF2 | 1e-101 | A0A096ZXF2_BETVU; R2R3-MYB flavonol regulator | ||||
| STRING | XP_010679349.1 | 1e-102 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G47460.1 | 2e-55 | myb domain protein 12 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AHYPO_008931-RA |



