 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AHYPO_008983-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Amaranthaceae; Amaranthus
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 340aa MW: 37775.2 Da PI: 6.5912 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 107.9 | 4.9e-34 | 173 | 230 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
+Dgy+WrKYGqK vk+s+fprsYYrCts++C+vkk+vers +dp++v++tYeg+H+h+
AHYPO_008983-RA 173 EDGYRWRKYGQKAVKNSPFPRSYYRCTSQSCNVKKRVERSFTDPSIVVTTYEGQHTHP 230
8********************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PIRSF | PIRSF038130 | 1.9E-96 | 2 | 339 | IPR017396 | PWRKY transcription factor, group IIc |
| Gene3D | G3DSA:2.20.25.80 | 2.6E-34 | 157 | 230 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 5.62E-29 | 164 | 230 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 31.043 | 167 | 232 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.2E-40 | 172 | 231 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 6.8E-27 | 173 | 230 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009624 | Biological Process | response to nematode | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 340 aa Download sequence Send to blast |
MEMKEPLKME TLKGGASLNT NNYHYLTENK CSSTTKSLDS QIVLENIYCP SSSTNNNNTG 60 FMELLGMQDM ININPPDLFD IMMMNGDDNQ AFHCSNVNLE SSEVINNNNL PTTPNSTSIS 120 SASNEALNDD QHQDDEQEQK TKKQCTKAKK TNEKRQREAR FAFMTKSEVD HLEDGYRWRK 180 YGQKAVKNSP FPRSYYRCTS QSCNVKKRVE RSFTDPSIVV TTYEGQHTHP CTVLPRGLIS 240 GSGYGSGLNA NIHGAISPFV VPMQMANNNH FQMNYGNSLV PDQLSQFGNV TIGSSVSPSC 300 SLLGTQERCN FGTPLQLNAA LLTDHGLLQD ILPSIMKKE* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 2e-27 | 165 | 229 | 10 | 74 | Probable WRKY transcription factor 4 |
| 2lex_A | 2e-27 | 165 | 229 | 10 | 74 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00069 | PBM | Transfer from AT2G47260 | Download |
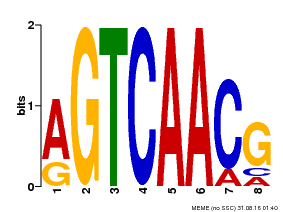
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | GU984040 | 5e-51 | GU984040.1 Cucumis sativus WRKY protein (WRKY55) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010679416.1 | 1e-126 | PREDICTED: probable WRKY transcription factor 23 | ||||
| Swissprot | O22900 | 1e-56 | WRK23_ARATH; WRKY transcription factor 23 | ||||
| TrEMBL | A0A0K9RLH1 | 1e-102 | A0A0K9RLH1_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010679416.1 | 1e-126 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G47260.1 | 3e-48 | WRKY DNA-binding protein 23 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AHYPO_008983-RA |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



