 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AHYPO_009144-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Amaranthaceae; Amaranthus
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 605aa MW: 69724.1 Da PI: 6.8535 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 112.6 | 6.3e-35 | 74 | 200 | 2 | 133 |
DUF822 2 gsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeaeaagssasaspesslq 95
g+gr+++++E+E++k+RER+RRai++++++GLR++Gn++lp+raD+n+Vl+AL+reAGw+ G + + +pl++++ ++s +as + +
AHYPO_009144-RA 74 GKGRREREREKERTKLRERHRRAITSRMLSGLRQYGNFPLPARADMNDVLAALAREAGWIG---GLPVKSSESPLSASSLKNRSMKASLDCQPT 164
789********************************************************95...888888889998888888888887776655 PP
DUF822 96 sslkssalaspvesysaspksssfpspssldsislasa 133
+ + ++s +++ ++ ++++ s+s+ ++
AHYPO_009144-RA 165 --VLGIDENLSIASLDSVMVTDMETKSEKYTSTSNIES 200
..444444444444444444444444444433333322 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 8.6E-31 | 74 | 133 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| SuperFamily | SSF51445 | 1.9E-140 | 210 | 605 | IPR017853 | Glycoside hydrolase superfamily |
| Gene3D | G3DSA:3.20.20.80 | 1.1E-146 | 210 | 535 | IPR013781 | Glycoside hydrolase, catalytic domain |
| Pfam | PF01373 | 2.8E-74 | 222 | 531 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.5E-46 | 235 | 249 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.5E-46 | 256 | 274 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.5E-46 | 278 | 299 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.5E-46 | 371 | 393 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.5E-46 | 444 | 463 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.5E-46 | 478 | 494 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.5E-46 | 495 | 506 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.5E-46 | 513 | 536 | IPR001554 | Glycoside hydrolase, family 14 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000272 | Biological Process | polysaccharide catabolic process | ||||
| GO:0048831 | Biological Process | regulation of shoot system development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0016161 | Molecular Function | beta-amylase activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 605 aa Download sequence Send to blast |
MNNAGKNTTP TTFNRIESQQ NPQNLTPQIP NPNPNNFSQS NSHHYQLISQ QQQNNRRPRG 60 FAVAANNSPA SGDGKGRRER EREKERTKLR ERHRRAITSR MLSGLRQYGN FPLPARADMN 120 DVLAALAREA GWIGGLPVKS SESPLSASSL KNRSMKASLD CQPTVLGIDE NLSIASLDSV 180 MVTDMETKSE KYTSTSNIES TDSLEAHQSG IINSYCQLLD PEGIRQELKH LKSLNVDGVV 240 VDCWWGIVEG WNPQKYMWLG YRELFTIIRE FQLKVQVVLA FHEYGGSDSG NVFISLPHWV 300 LEIGKDNQEI FFTDREGRRN TECLSWGIDK ERVLRGRTGI EVYFDFMRSF RSEFDDIFCE 360 GLITAVEIGL GPSGDLKYPS FPERLGWRYP GIGEFQCYDR YMQQNLRKAA KLRGHSYWAR 420 GPDNAGNYNS KPSETGFFRE RGDYDSYFGR FFLHWYAQML IDHADNVLSL ARLAFDETQI 480 VVKIPAVHWW YKTPSHAAEL TAGFYNPTNR DGYSPVFDVL RKHSVTVKLV CPRSHVHFRD 540 NDESFADPED PCFDREEHLR MIETSKPRDD PDHHHFSFFQ YQLPPPFVQR EYCFSELDYF 600 IKCMH |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wdq_A | 1e-108 | 210 | 570 | 21 | 374 | Beta-amylase |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00543 | DAP | Transfer from AT5G45300 | Download |
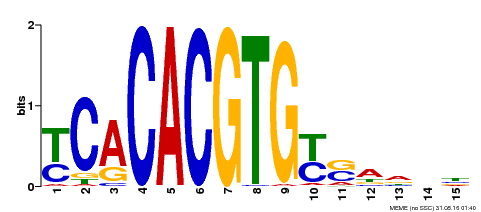
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021858576.1 | 0.0 | beta-amylase 8 | ||||
| Swissprot | Q9FH80 | 0.0 | BAM8_ARATH; Beta-amylase 8 | ||||
| TrEMBL | A0A0K9QGA3 | 0.0 | A0A0K9QGA3_SPIOL; Beta-amylase | ||||
| STRING | XP_010676684.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45300.1 | 0.0 | beta-amylase 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AHYPO_009144-RA |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



