 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AHYPO_011703-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Amaranthaceae; Amaranthus
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 477aa MW: 50637.3 Da PI: 10.0374 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 50.1 | 5.8e-16 | 399 | 449 | 5 | 55 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkke 55
+r+rr++kNRe+A rsR+RK+a++ eLe++v++L++eN++L+k+ e+ ++
AHYPO_011703-RA 399 RRQRRMIKNRESAARSRARKQAYTMELEQEVQKLKEENQDLRKKQAEIIEM 449
79****************************************987776555 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF81995 | 2.88E-7 | 239 | 294 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 5.8E-15 | 393 | 446 | No hit | No description |
| SMART | SM00338 | 1.2E-14 | 395 | 460 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 11.599 | 397 | 446 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 4.88E-11 | 399 | 447 | No hit | No description |
| CDD | cd14707 | 8.10E-26 | 399 | 453 | No hit | No description |
| Pfam | PF00170 | 4.1E-14 | 399 | 449 | IPR004827 | Basic-leucine zipper domain |
| PROSITE pattern | PS00036 | 0 | 402 | 417 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009738 | Biological Process | abscisic acid-activated signaling pathway | ||||
| GO:0010255 | Biological Process | glucose mediated signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 477 aa Download sequence Send to blast |
MGSNCNINFS DGGAGGSGRP PGSGNFGLTR QSSIYSLTFD ELQNTLGGMG KDFGSMNMDE 60 LLKNIWSAEE TQSMATGSGA AAMAAAAAAV ANMGQDVGNS GGYLQRQGSL TLPRTLSQKT 120 VDEVWKDIAK EFNGGKDGGG GSNVPQRQQT LGEITLEEFL VRAGVVREDA QVVGKPNNVP 180 GVGIFGDFAQ PDHNHSNSNS GFGINFQQPG RGMDLIGNRI SETNHQMASL QSANLPLNVN 240 GVRTSQQQAA QVQLSQQQQQ QQLQQQLQQQ QQQQQQQQQQ QPLFPKQSTV SYGSAITLSN 300 SSPLGNAGMR GGMVGLADPG LNSNLVQSGA NQGGAGLGMV GLGGGGTIGV PAGSPAALSS 360 DGIARSNGDS SSVSPVPYMF NGGLRGRRGL GTVDKVVERR QRRMIKNRES AARSRARKQA 420 YTMELEQEVQ KLKEENQDLR KKQAEIIEMQ KNQVMEMMNA QSGPRKKLRR TQTGPW* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 133 | 140 | GGKDGGGG |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Involved in ABA and stress responses and acts as a positive component of glucose signal transduction. Functions as transcriptional activator in the ABA-inducible expression of rd29B. Binds specifically to the ABA-responsive element (ABRE) of the rd29B gene promoter. {ECO:0000269|PubMed:11005831, ECO:0000269|PubMed:15361142, ECO:0000269|PubMed:16284313, ECO:0000269|PubMed:16463099}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00186 | DAP | Transfer from AT1G45249 | Download |
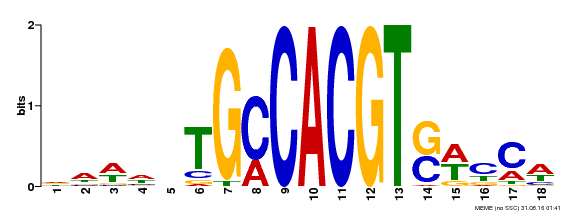
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by drought, salt, abscisic acid (ABA), cold and glucose. {ECO:0000269|PubMed:10636868, ECO:0000269|PubMed:11005831, ECO:0000269|PubMed:15361142, ECO:0000269|PubMed:16284313, ECO:0000269|PubMed:16463099}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021716588.1 | 0.0 | ABSCISIC ACID-INSENSITIVE 5-like protein 5 | ||||
| Swissprot | Q9M7Q4 | 1e-123 | AI5L5_ARATH; ABSCISIC ACID-INSENSITIVE 5-like protein 5 | ||||
| TrEMBL | B5QTD2 | 0.0 | B5QTD2_BETVU; ABA-responsive element binding protein 1 | ||||
| STRING | XP_010685236.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G45249.1 | 1e-100 | abscisic acid responsive elements-binding factor 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AHYPO_011703-RA |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



