 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AHYPO_012412-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Amaranthaceae; Amaranthus
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 338aa MW: 37734.2 Da PI: 9.6759 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 101.4 | 5.8e-32 | 80 | 133 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
prlrWtp+LH +F++ave+LGG+e+AtPk +l+lm+vkgL+++hvkSHLQ+YR+
AHYPO_012412-RA 80 PRLRWTPDLHLCFIQAVERLGGQERATPKLVLQLMNVKGLSIAHVKSHLQMYRS 133
8****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 2.8E-30 | 76 | 134 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 13.346 | 76 | 136 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.2E-14 | 78 | 134 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 9.8E-24 | 80 | 135 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 1.3E-9 | 81 | 132 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 338 aa Download sequence Send to blast |
MIRARSGSTL EGSKSSLSNS KIDDDSIEEV DDNSNHSNSN SKLLKVNGSS SSSSTVEESD 60 KKTLSSSSGS VRPYNRSKMP RLRWTPDLHL CFIQAVERLG GQERATPKLV LQLMNVKGLS 120 IAHVKSHLQM YRSKKIDESN QAMSNQGVFM DGRDNIYDLS QLPMLQAYNN QRSSFNLSNS 180 QANVHDFQGK FPNWITQQTL KKDPMSQTNE SPTSKSSMEF DLAIRSISGS AEGREKSNSL 240 NFLENDSKIE KNQKETFSHL KRKAMDLDLN LTLASKDTIF NGLTKKFAKH NEVLGSELTL 300 SLQHPSPLFS SRLDQDKFIS KFKEGNEQAR TSLDLTL* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 8e-19 | 81 | 135 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 8e-19 | 81 | 135 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_A | 9e-19 | 81 | 135 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 9e-19 | 81 | 135 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 9e-19 | 81 | 135 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 9e-19 | 81 | 135 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 8e-19 | 81 | 135 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 8e-19 | 81 | 135 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 8e-19 | 81 | 135 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 8e-19 | 81 | 135 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 8e-19 | 81 | 135 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 8e-19 | 81 | 135 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00297 | DAP | Transfer from AT2G38300 | Download |
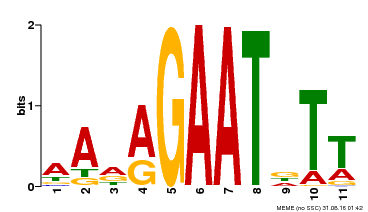
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021732658.1 | 1e-126 | two-component response regulator ARR1-like | ||||
| TrEMBL | A0A0J8BEJ5 | 1e-105 | A0A0J8BEJ5_BETVU; Uncharacterized protein | ||||
| STRING | XP_010693076.1 | 1e-106 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G40260.1 | 6e-47 | G2-like family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AHYPO_012412-RA |



