 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AHYPO_012536-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Amaranthaceae; Amaranthus
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 152aa MW: 16915 Da PI: 9.6558 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 55.5 | 1.4e-17 | 15 | 64 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
ykG+r +k +g+WvAeIr+p +kr r++lg++ t+ Aa+a++ a +l+g
AHYPO_012536-RA 15 PYKGIRMRK-WGKWVAEIREP---NKRSRIWLGSYSTPVAAARAYDTAVYYLRG 64
69*****99.**********9...336*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51032 | 21.904 | 15 | 72 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 5.17E-21 | 15 | 73 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 6.5E-35 | 15 | 78 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 6.6E-30 | 16 | 73 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 2.8E-11 | 16 | 27 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 1.37E-26 | 16 | 74 | No hit | No description |
| Pfam | PF00847 | 7.0E-11 | 16 | 64 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 2.8E-11 | 54 | 74 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 152 aa Download sequence Send to blast |
MERGSTKKRG RAEKPYKGIR MRKWGKWVAE IREPNKRSRI WLGSYSTPVA AARAYDTAVY 60 YLRGPSARLN FPDLLIGDCR LSDLSAASIR KKAIEVGARV DALQTTFQPP PTTNTNSGKV 120 SEPEDAGNNT GFVDEKPDLN KLPEDSDEWE D* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5wx9_A | 8e-15 | 10 | 72 | 9 | 72 | Ethylene-responsive transcription factor ERF096 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). {ECO:0000250}. | |||||
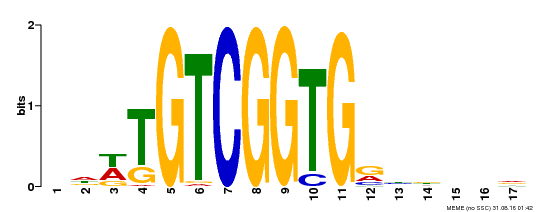
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00276 | DAP | Transfer from AT2G23340 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021714345.1 | 4e-78 | ethylene-responsive transcription factor ERF010-like | ||||
| Swissprot | O22174 | 9e-51 | ERF08_ARATH; Ethylene-responsive transcription factor ERF008 | ||||
| TrEMBL | A0A0K9QM16 | 3e-73 | A0A0K9QM16_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010679510.1 | 1e-72 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G23340.1 | 3e-50 | DREB and EAR motif protein 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AHYPO_012536-RA |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



