 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AHYPO_012588-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Amaranthaceae; Amaranthus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 293aa MW: 32937.8 Da PI: 6.5136 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 56.4 | 7e-18 | 14 | 61 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEd +lv +++++G+g+W++++ g++R+ k+c++rw +yl
AHYPO_012588-RA 14 KGPWTPEEDIILVSYIQEHGPGNWRAVPTNTGLRRCSKSCRLRWTNYL 61
79********************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 49.1 | 1.3e-15 | 67 | 112 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg++T+ E++ ++++ ++lG++ W++Ia++++ Rt++++k++w+++l
AHYPO_012588-RA 67 RGNFTEKEEKMIIHLQALLGNR-WAAIASYLP-QRTDNDIKNYWNTHL 112
89********************.*********.************996 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 2.3E-24 | 5 | 64 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 25.663 | 9 | 65 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 7.19E-31 | 11 | 108 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 8.3E-15 | 13 | 63 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.4E-16 | 14 | 61 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 6.41E-11 | 16 | 61 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 8.4E-25 | 65 | 117 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 20.464 | 66 | 116 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.9E-15 | 66 | 114 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 8.0E-15 | 67 | 112 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.08E-10 | 69 | 112 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0080167 | Biological Process | response to karrikin | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 293 aa Download sequence Send to blast |
MGRPPCCDKI GVKKGPWTPE EDIILVSYIQ EHGPGNWRAV PTNTGLRRCS KSCRLRWTNY 60 LRPGIKRGNF TEKEEKMIIH LQALLGNRWA AIASYLPQRT DNDIKNYWNT HLKKKIKKLE 120 EGQDGQNTSS LIGFSSNSTS TSSHNSNKGQ WERRLQTDIH MAKQALCEAL SIEKPELHPS 180 PSPIYASSAE NIARLLENWM TRKPNSGVSS SSSPQESMTQ NFNEELKPVQ SLYSNSNSST 240 TTDDSHESVS AEAEAAQQVP LSLLEKWLFE EGVNMMDMSL DVDHETDQVD LF* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gv2_A | 5e-26 | 14 | 116 | 4 | 105 | MYB PROTO-ONCOGENE PROTEIN |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator involved in the activation of cuticular wax biosynthesis under drought stress. Binds directly to the promoters of genes involved in cuticular wax biosynthesis. Transactivates WSD1, KCS2/DAISY, CER1, CER2, FAR3 and ECR genes (PubMed:25305760, PubMed:27577115). Functions together with MYB96 in the activation of cuticular wax biosynthesis (PubMed:27577115). {ECO:0000269|PubMed:25305760, ECO:0000269|PubMed:27577115}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00395 | DAP | Transfer from AT3G47600 | Download |
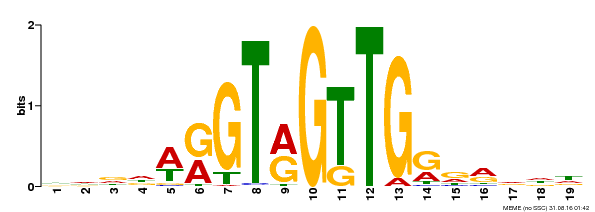
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by salt stress, osmotic shock and abscisic acid (ABA). {ECO:0000269|PubMed:25305760}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021860329.1 | 1e-138 | myb-related protein 306-like | ||||
| Swissprot | Q9SN78 | 1e-117 | MYB94_ARATH; Transcription factor MYB94 | ||||
| TrEMBL | A0A2N9J4F3 | 1e-121 | A0A2N9J4F3_FAGSY; Uncharacterized protein | ||||
| STRING | XP_010678525.1 | 1e-131 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G47600.1 | 1e-108 | myb domain protein 94 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AHYPO_012588-RA |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



