 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AHYPO_012667-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Amaranthaceae; Amaranthus
|
||||||||
| Family | SRS | ||||||||
| Protein Properties | Length: 307aa MW: 33378.6 Da PI: 6.332 | ||||||||
| Description | SRS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF702 | 211.4 | 2.2e-65 | 67 | 227 | 3 | 153 |
DUF702 3 sgtasCqdCGnqakkdCaheRCRtCCksrgfdCathvkstWvpaakrrerqqqlaaasskaaasa.....aeaaskrkrelkskkqsalsstkl 91
g++sCqdCGnqakkdC+++RCRtCCksrgfdCathv+stWvpa+ rr+r+q+l + ++++++ +skr+ + +++s+l +t+l
AHYPO_012667-RA 67 GGSISCQDCGNQAKKDCVYMRCRTCCKSRGFDCATHVRSTWVPASVRRDRHQKLVQHLQHQQEQDqeqmgIGLNSKRR---RDHSSSSLVCTQL 157
5789**********************************************7776655554444442433312223333...3456667777777 PP
DUF702 92 ssaeskkeletss........lPeevsseavfrcvrvssvddgeeelaYqtavsigGhvfkGiLydqGle 153
+ ++++++++ s +P+e+s +a frcv+vss+dd+e+e+aYqtav+igGhvfkGiLydqG+
AHYPO_012667-RA 158 PASDTNTNTTSSFsgpmevgnFPAELSYPATFRCVKVSSIDDSEDEYAYQTAVNIGGHVFKGILYDQGPG 227
766655444333356777777***********************************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05142 | 1.3E-64 | 69 | 226 | IPR007818 | Protein of unknown function DUF702 |
| TIGRFAMs | TIGR01623 | 1.3E-27 | 71 | 112 | IPR006510 | Zinc finger, lateral root primordium type 1 |
| TIGRFAMs | TIGR01624 | 1.3E-26 | 179 | 226 | IPR006511 | Lateral Root Primordium type 1, C-terminal |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010051 | Biological Process | xylem and phloem pattern formation | ||||
| GO:0010252 | Biological Process | auxin homeostasis | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0048479 | Biological Process | style development | ||||
| GO:0048480 | Biological Process | stigma development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0046982 | Molecular Function | protein heterodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 307 aa Download sequence Send to blast |
MAGFFSLDHN TNTNNSYLWT NHHHHHQEPP LNLSISAVDF EGGNDITRSE DNSNNFMMMR 60 MSSSSSGGSI SCQDCGNQAK KDCVYMRCRT CCKSRGFDCA THVRSTWVPA SVRRDRHQKL 120 VQHLQHQQEQ DQEQMGIGLN SKRRRDHSSS SLVCTQLPAS DTNTNTTSSF SGPMEVGNFP 180 AELSYPATFR CVKVSSIDDS EDEYAYQTAV NIGGHVFKGI LYDQGPGDHH DHPQPGDAGG 240 SGGGETPSGG GNLTMGPTTS VSGTLFEMPT SSSSPVVGYL DPSSYYPLPL NAYLPPGTPF 300 FPYPRP* |
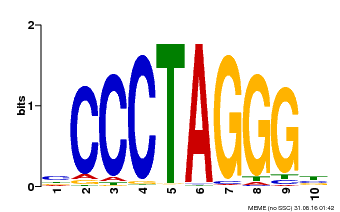
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00613 | PBM | Transfer from AT3G51060 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021718103.1 | 1e-108 | protein SHI RELATED SEQUENCE 1-like isoform X1 | ||||
| TrEMBL | A0A0K9QJA8 | 9e-81 | A0A0K9QJA8_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010679824.1 | 6e-86 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G51060.1 | 4e-53 | Lateral root primordium (LRP) protein-related | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AHYPO_012667-RA |



