 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AHYPO_013438-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Amaranthaceae; Amaranthus
|
||||||||
| Family | TALE | ||||||||
| Protein Properties | Length: 313aa MW: 35890.3 Da PI: 4.8599 | ||||||||
| Description | TALE family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 27.7 | 4.5e-09 | 255 | 289 | 21 | 55 |
HSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHH CS
Homeobox 21 knrypsaeereeLAkklgLterqVkvWFqNrRake 55
k +yp+++++ LA+ +gL++rq+ +WF N+R ++
AHYPO_013438-RA 255 KWPYPTEADKIALAEMTGLDQRQINNWFINQRKRH 289
569*****************************885 PP
| |||||||
| 2 | ELK | 30.3 | 9.7e-11 | 209 | 230 | 1 | 22 |
ELK 1 ELKhqLlrKYsgyLgsLkqEFs 22
+LK+qLlr ++++++sLk EFs
AHYPO_013438-RA 209 DLKNQLLRRFGSHISSLKLEFS 230
6********************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01255 | 9.3E-21 | 72 | 116 | IPR005540 | KNOX1 |
| Pfam | PF03790 | 4.4E-22 | 74 | 115 | IPR005540 | KNOX1 |
| Pfam | PF03789 | 1.8E-8 | 209 | 230 | IPR005539 | ELK domain |
| SMART | SM01188 | 2.6E-5 | 209 | 230 | IPR005539 | ELK domain |
| PROSITE profile | PS51213 | 9.725 | 209 | 229 | IPR005539 | ELK domain |
| PROSITE profile | PS50071 | 12.73 | 229 | 292 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 2.74E-20 | 230 | 304 | IPR009057 | Homeodomain-like |
| SMART | SM00389 | 3.8E-13 | 231 | 296 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 3.9E-28 | 234 | 294 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 3.44E-13 | 238 | 293 | No hit | No description |
| Pfam | PF05920 | 8.1E-18 | 249 | 288 | IPR008422 | Homeobox KN domain |
| PROSITE pattern | PS00027 | 0 | 267 | 290 | IPR017970 | Homeobox, conserved site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010073 | Biological Process | meristem maintenance | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 313 aa Download sequence Send to blast |
MDDMYGIHHQ TVEDYSDKSL ISPDNLILPS DYHSLIMNSG NTTAVRFGSD ELINATCLSE 60 STSVTPDDHT SNSIKIKIAS HPHYPRLLQA YIDCHKVGAP PEIANILDEI KQESDMYSQD 120 IGRGRRTCLG MDPELDEFMD YLLVIIRNKG MLITLWVVIF RRYGDSETIR ESSQEQGSIE 180 EEGVVSSDED CSGGEIDVQD LQPKDGEQDL KNQLLRRFGS HISSLKLEFS KKKKKGKLPR 240 EARQMLFEWW NAHYKWPYPT EADKIALAEM TGLDQRQINN WFINQRKRHW KPSENMQYAL 300 MDGFAGQYFT DD* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Plays a role in meristem function. Contributes to the shoot apical meristem (SAM) maintenance and organ separation by controlling boundary establishment in embryo in a CUC1, CUC2 and STM-dependent manner. Involved in maintaining cells in an undifferentiated, meristematic state. Probably binds to the DNA sequence 5'-TGAC-3'. {ECO:0000269|PubMed:16798887}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00675 | ChIP-seq | Transfer from GRMZM2G017087 | Download |
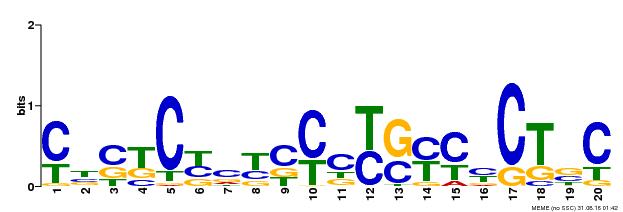
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Seems to be repressed by AS2 and AS1 but induced by STM, CUC1 and CUC2. {ECO:0000269|PubMed:11311158, ECO:0000269|PubMed:16798887}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | DQ374017 | 6e-69 | DQ374017.1 Beta vulgaris chromosome 9 clone BAC62 genomic sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010695813.1 | 1e-173 | PREDICTED: homeobox protein knotted-1-like 6 isoform X1 | ||||
| Swissprot | Q84JS6 | 1e-105 | KNAT6_ARATH; Homeobox protein knotted-1-like 6 | ||||
| TrEMBL | A0A0J8BBK5 | 1e-171 | A0A0J8BBK5_BETVU; Uncharacterized protein | ||||
| STRING | XP_010695813.1 | 1e-172 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G23380.1 | 6e-92 | KNOTTED1-like homeobox gene 6 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AHYPO_013438-RA |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



