 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AHYPO_014407-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Amaranthaceae; Amaranthus
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 244aa MW: 28415.2 Da PI: 9.9602 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 100.3 | 1.2e-31 | 187 | 244 | 1 | 58 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnh 58
ldDgy+WrKYGqK+vk+s +prsYYrCt+++C+vkk+ver +ed ++v++tYeg+Hnh
AHYPO_014407-RA 187 LDDGYKWRKYGQKVVKNSLHPRSYYRCTHNNCRVKKRVERLSEDCRMVITTYEGRHNH 244
59*******************************************************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 3.9E-33 | 172 | 244 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 4.18E-28 | 179 | 244 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 30.274 | 182 | 244 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.3E-33 | 187 | 244 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 7.6E-25 | 188 | 244 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 244 aa Download sequence Send to blast |
MERDHTKNNN NFLNYDLHMS LIHTNNHHQE NSSNNINLQG FHTLELPNNH HPTNNNLMLT 60 TFLHPPPPQP PPSTYHQITR HRTLTDAPNT ATTTTTTKGL VGNFDNYVAS TRQSWNSNNT 120 HHQQVEGSSD PKATVNGDHS NGNGRNDNGA DHSWWKSSNS EKNKMKIRRK LREPRFCFQT 180 RSDIDVLDDG YKWRKYGQKV VKNSLHPRSY YRCTHNNCRV KKRVERLSED CRMVITTYEG 240 RHNH |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 8e-26 | 178 | 244 | 8 | 74 | Probable WRKY transcription factor 4 |
| 2lex_A | 8e-26 | 178 | 244 | 8 | 74 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 165 | 171 | KIRRKLR |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00618 | PBM | Transfer from AT2G44745 | Download |
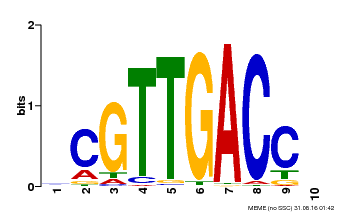
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010692038.1 | 1e-113 | PREDICTED: probable WRKY transcription factor 48 | ||||
| TrEMBL | A0A0K9RFP6 | 3e-70 | A0A0K9RFP6_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010692038.1 | 1e-113 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G44745.1 | 2e-54 | WRKY family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AHYPO_014407-RA |



