 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AHYPO_014422-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Amaranthaceae; Amaranthus
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 457aa MW: 50072.7 Da PI: 6.5648 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 74.1 | 2e-23 | 255 | 301 | 1 | 47 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvk 47
k+r++W+p+LH+rFv+a++ LGGs++AtPk+i+elmkv+gLt+++vk
AHYPO_014422-RA 255 KARRCWSPDLHRRFVNALQMLGGSQVATPKQIRELMKVDGLTNDEVK 301
68********************************************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 8.24E-9 | 252 | 301 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 2.2E-18 | 253 | 301 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 4.7E-18 | 255 | 301 | IPR006447 | Myb domain, plants |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009266 | Biological Process | response to temperature stimulus | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010452 | Biological Process | histone H3-K36 methylation | ||||
| GO:0048579 | Biological Process | negative regulation of long-day photoperiodism, flowering | ||||
| GO:1903507 | Biological Process | negative regulation of nucleic acid-templated transcription | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 457 aa Download sequence Send to blast |
MMTSPTELSL ECKPQSYSML LKSFGGDVIN NPTDQQATQK LQDYLTRLEE ERLKIDAFKR 60 ELPLCMQLLT QAMEVSRQQL QSCRSNNEGT RPVLEEFMPI KQSNMEGSDQ KQTNTSDKEN 120 WMLSAQLWSQ AGDPNKQQSS ILPKSEADHN TFNNVGNCPK LGLDNKQRNG GAFLPFTKER 180 SLIPNNSSIM ALPDLALVSN EREMEEKKCT DSENGMSDNS NKISNVMVGK AVDSQSPTNT 240 TTTPATPTSS QTHRKARRCW SPDLHRRFVN ALQMLGGSQV ATPKQIRELM KVDGLTNDEV 300 KRAPAPQLVV LGGIWVPPEY ATAAAAAHGG PPTIYSTHPS TQVSSHYCSP TVPSDFYPQA 360 SSAPQALHLS ALHHHQHPQL HVYKTASHTQ TTHSSPESDG HGAGNRSESM EDGKSESSSW 420 KGESGGENGG DQRNKLAALR VEDGESMNGS EITLKF* |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00261 | DAP | Transfer from AT2G03500 | Download |
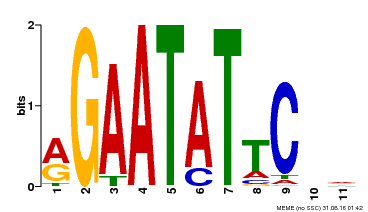
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021866139.1 | 0.0 | myb family transcription factor EFM | ||||
| TrEMBL | A0A0K9RN43 | 0.0 | A0A0K9RN43_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010688115.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G03500.1 | 1e-102 | G2-like family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AHYPO_014422-RA |



