 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AHYPO_014705-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Amaranthaceae; Amaranthus
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 509aa MW: 55626.4 Da PI: 7.1439 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 107.2 | 8.3e-34 | 223 | 279 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
dDgynWrKYGqK+vkgsefprsYY+Ct+a+Cpv+kkvers d++v+ei+Y+g+Hnh+
AHYPO_014705-RA 223 DDGYNWRKYGQKHVKGSEFPRSYYKCTHASCPVRKKVERSL-DGQVTEIIYKGQHNHQ 279
8****************************************.***************8 PP
| |||||||
| 2 | WRKY | 88.1 | 7.5e-28 | 397 | 464 | 1 | 59 |
---SS-EEEEEEE--TT-SS-E.........EEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefpr.........sYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+vkg+++pr sYY+Ct++gC+v+k+ver+ +dpk+v++tYeg+Hnh+
AHYPO_014705-RA 397 LDDGYRWRKYGQKVVKGNPYPRqfnvaaaftSYYKCTHPGCNVRKQVERAPSDPKAVVTTYEGKHNHD 464
59*******************97777777778***********************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 1.9E-30 | 208 | 281 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 2.62E-26 | 220 | 281 | IPR003657 | WRKY domain |
| SMART | SM00774 | 2.6E-37 | 222 | 280 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 24.248 | 223 | 281 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 6.3E-26 | 223 | 279 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 6.9E-34 | 382 | 466 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.44E-26 | 389 | 466 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 32.994 | 392 | 466 | IPR003657 | WRKY domain |
| SMART | SM00774 | 2.5E-38 | 397 | 465 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 8.9E-21 | 398 | 464 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 509 aa Download sequence Send to blast |
MAENDPIPST KPPQPQRPTI TLPSRAGIES LFSGSGFGLS PGPMTLVSSF FSENDPDSDC 60 RSFSQLLAGA IASPRPPSEP DQLRFKDNRP PGLVISQPQP QSQVGVGVGG GFFLLSPGVS 120 PALLLDSPGL FPGQGPFGMS HQQALAQVTA QAQAMQQAQQ GGFQSSTTVG GSEPQSTFTP 180 ELEMQQKMMS SASESKETEE PAQYSGSEQR LQTSSIIVDK PADDGYNWRK YGQKHVKGSE 240 FPRSYYKCTH ASCPVRKKVE RSLDGQVTEI IYKGQHNHQP PQKKSKDGGN PNGNLGNQGN 300 PELVSQTQPN NTNSMSEGMA RSLSRRDQES SQATADQLSS DEEEVDNDRT KEAGGDEDER 360 DAKRRNIEVR VTEQAASHRT VTEPKIVVQT TSEVDLLDDG YRWRKYGQKV VKGNPYPRQF 420 NVAAAFTSYY KCTHPGCNVR KQVERAPSDP KAVVTTYEGK HNHDIPAARM SSHNTANSSS 480 HLRPHKTEYA NTREQPVATL RLKEERIT* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 1e-38 | 223 | 467 | 8 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 1e-38 | 223 | 467 | 8 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00260 | DAP | Transfer from AT2G03340 | Download |
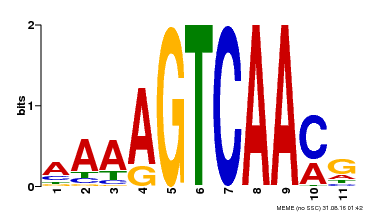
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salicylic acid and during leaf senescence. {ECO:0000269|PubMed:11449049, ECO:0000269|PubMed:11722756}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021739996.1 | 0.0 | probable WRKY transcription factor 3 | ||||
| Swissprot | Q9ZQ70 | 1e-155 | WRKY3_ARATH; Probable WRKY transcription factor 3 | ||||
| TrEMBL | A0A0K9R3N8 | 0.0 | A0A0K9R3N8_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010683088.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G03340.1 | 1e-122 | WRKY DNA-binding protein 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AHYPO_014705-RA |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



