 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AHYPO_016595-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Amaranthaceae; Amaranthus
|
||||||||
| Family | TCP | ||||||||
| Protein Properties | Length: 471aa MW: 50986.9 Da PI: 9.6088 | ||||||||
| Description | TCP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | TCP | 119.4 | 4.2e-37 | 81 | 182 | 1 | 89 |
TCP 1 aagkkdrhskihTkvggRdRRvRlsaecaarfFdLqdeLGfdkdsktieWLlqqakpaikeltgtssssaseceaesssssasnsssg...... 88
a+g+kdrhsk++T++g+RdRRvRlsa++a++f+d+qd+LG+d++sk+++WL+++ak ai+el ++++ s+s+++ ++s+ +++ +
AHYPO_016595-RA 81 ATGRKDRHSKVCTAKGPRDRRVRLSAHTAIQFYDVQDRLGYDRPSKAVDWLIKKAKNAIDELEQLPAWSPSSMNMSTSNRRRKKNDVKddmqee 174
589**************************************************************77777555222222222211111223333 PP
TCP 89 .......k 89
+
AHYPO_016595-RA 175 kgggsggS 182
34554330 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03634 | 9.8E-34 | 82 | 191 | IPR005333 | Transcription factor, TCP |
| PROSITE profile | PS51369 | 34.001 | 84 | 142 | IPR017887 | Transcription factor TCP subgroup |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009793 | Biological Process | embryo development ending in seed dormancy | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0030154 | Biological Process | cell differentiation | ||||
| GO:0045962 | Biological Process | positive regulation of development, heterochronic | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 471 aa Download sequence Send to blast |
MKGNHFHHYN CKNNHHSIII KSEQPIKKRA YFPSSSSSSI PAGTEQVQQQ QKKQPGSSRT 60 SLRKAGIGEI VEVQGGHIVR ATGRKDRHSK VCTAKGPRDR RVRLSAHTAI QFYDVQDRLG 120 YDRPSKAVDW LIKKAKNAID ELEQLPAWSP SSMNMSTSNR RRKKNDVKDD MQEEKGGGSG 180 GSGERLYGGE DQQQLYNSSN VNNNSGSSLL PPSLDSDVIA DTMKSFFPMG VGGNVGPTSN 240 SSASEFQDYF HGGLLQRSGG NGRQNQDLKL SLQSFQDPIL MHSHSHHGGS AAHNEQQQAL 300 FPGAVPIGFE NPHTDWGDSQ HHPSPELGRL HRLVVGWNTQ QTGTEHHSVG TGSGSSNGGG 360 SGYVFNQPSP LLQPLFGGGQ HSQFLSQRGP LQSSNTPSIR AWLSDPSFSI SSSSEHHNHQ 420 AIQVHPQALS GLGFVSGGFS GFHIPARIQG EEEQNGDLNR PSSASSGSRH * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5zkt_A | 1e-20 | 89 | 142 | 1 | 54 | Putative transcription factor PCF6 |
| 5zkt_B | 1e-20 | 89 | 142 | 1 | 54 | Putative transcription factor PCF6 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor playing a pivotal role in the control of morphogenesis of shoot organs by negatively regulating the expression of boundary-specific genes such as CUC genes, probably through the induction of miRNA (e.g. miR164) (PubMed:12931144, PubMed:17307931). Required during early steps of embryogenesis (PubMed:15634699). Participates in ovule develpment (PubMed:25378179). Activates LOX2 expression by binding to the 5'-GGACCA-3' motif found in its promoter (PubMed:18816164). {ECO:0000269|PubMed:12931144, ECO:0000269|PubMed:15634699, ECO:0000269|PubMed:17307931, ECO:0000269|PubMed:18816164, ECO:0000269|PubMed:25378179}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00009 | PBM | Transfer from 929286 | Download |
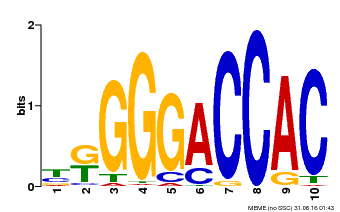
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by the miRNA miR-JAW/miR319 (PubMed:12931144, PubMed:18816164). {ECO:0000269|PubMed:12931144, ECO:0000269|PubMed:18816164}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021743137.1 | 0.0 | LOW QUALITY PROTEIN: transcription factor TCP4-like | ||||
| Swissprot | Q8LPR5 | 6e-93 | TCP4_ARATH; Transcription factor TCP4 | ||||
| TrEMBL | A0A0K9RU33 | 0.0 | A0A0K9RU33_SPIOL; Uncharacterized protein | ||||
| STRING | XP_004137216.1 | 1e-121 | (Cucumis sativus) | ||||
| STRING | XP_004172687.1 | 1e-121 | (Cucumis sativus) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G15030.3 | 2e-47 | TCP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AHYPO_016595-RA |



