 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AHYPO_018354-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Amaranthaceae; Amaranthus
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 658aa MW: 74146.5 Da PI: 7.4411 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 34.9 | 3.6e-11 | 604 | 652 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHT...TTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmg...kgRtlkqcksrwqky 47
r +WT++E++ l d+vk++G Wk I ++ Rt ++k++w++
AHYPO_018354-RA 604 RRKWTPQEEDTLRDGVKRFGRS-WKYILECYReifGDRTYVDLKDKWRNM 652
679*****************55.*************************95 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 12.401 | 599 | 657 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.0E-12 | 601 | 654 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.7E-6 | 603 | 655 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.4E-8 | 604 | 652 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 7.0E-11 | 604 | 654 | IPR009057 | Homeodomain-like |
| CDD | cd11660 | 4.53E-16 | 605 | 654 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009901 | Biological Process | anther dehiscence | ||||
| GO:0010152 | Biological Process | pollen maturation | ||||
| GO:0043067 | Biological Process | regulation of programmed cell death | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 658 aa Download sequence Send to blast |
MDEKVAKWII EFLLRKTFID EKIINGVLLC LPISNNDSRL KKTILLRKIE SEISNGSVSE 60 NLLELFEIIE ELDHREGTRA SDSMKKAYCA VAVDCTLRFL EENAKDNRSY VLAVERVWRN 120 RVRLMEALVS EESKCWLEDI EAAIWDSEVC EKILMRNTRN EALKAVRGFL KEAWNSMGPA 180 FLELVAAKVG DEVEALRHGF GSVNHCRKVM ASQGVQKGQS LVRRKDVAVK TKHLRGAKIS 240 DAGEDEVDSI NTANDVPIDE IEKVPETPQS NSLQLQADAG GFHPDTSQVA QNLSFTKEND 300 VPTHNIDSVL APQLNSNVRA CHSDVCQDTH NMSHAKDNEL LPRTIEKLQA HQSSSMLSDS 360 HPDGSRAACT KFVTKVTDNA NGVKAAENRT DPVIQSPKVT RFQKALKTSV MELHEAPTEP 420 LQETIQVAET LCSTGVTHNH KTAVQNQTRV EKYAFTGVNR VQRALETSSV EVHAAGKNPG 480 ANASRTVENV SPKVATNNVK IDAVLVNQTQ PGTDAPRESP KNNPKNVDKD QTLEKIEGSD 540 THYPRELPKR KRSLMEPNST SHTMEWDDIE SRDLHAHSPS ISKRSRSSAL KLYDSRNFAI 600 RRMRRKWTPQ EEDTLRDGVK RFGRSWKYIL ECYREIFGDR TYVDLKDKWR NMTRYES* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 595 | 604 | RNFAIRRMRR |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00052 | PBM | Transfer from AT1G15720 | Download |
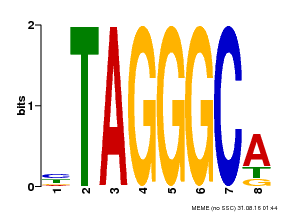
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021734347.1 | 0.0 | uncharacterized protein LOC110701068 | ||||
| TrEMBL | A0A0K9QSX4 | 1e-174 | A0A0K9QSX4_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010673889.1 | 1e-180 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G15720.1 | 5e-19 | TRF-like 5 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AHYPO_018354-RA |



