 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AHYPO_018564-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Amaranthaceae; Amaranthus
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 302aa MW: 33470.3 Da PI: 5.7392 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 62.5 | 8.9e-20 | 84 | 134 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
++y+GVr+++ +g+W+AeIrdp r r +lg+++taeeAa +++a+ +l+g
AHYPO_018564-RA 84 KKYRGVRQRP-WGKWAAEIRDPL----RrVRLWLGTYDTAEEAAMVYDNAAIQLRG 134
59********.**********72....45*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 8.44E-33 | 84 | 141 | No hit | No description |
| Pfam | PF00847 | 5.3E-13 | 85 | 134 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 5.6E-36 | 85 | 148 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 1.6E-30 | 85 | 142 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 3.73E-22 | 85 | 142 | IPR016177 | DNA-binding domain |
| PROSITE profile | PS51032 | 23.749 | 85 | 142 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 9.3E-11 | 86 | 97 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 9.3E-11 | 108 | 124 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0042991 | Biological Process | transcription factor import into nucleus | ||||
| GO:0048364 | Biological Process | root development | ||||
| GO:0048825 | Biological Process | cotyledon development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 302 aa Download sequence Send to blast |
MSFPKVVRIS VTDADATDSS SDDEVCIQRR RVKRFVNEVN IEPCSSCSAT TRVVDDVVLK 60 RRQRNKKKGK CSSVKQVQVN KSEKKYRGVR QRPWGKWAAE IRDPLRRVRL WLGTYDTAEE 120 AAMVYDNAAI QLRGPHALTN FSPPTKLFLS STSSSEDGSL SHPHHPSSIC SPTSVLRFQP 180 ESEPTSPTEA RKAPVPRPIK EEKKDEELVS GPTTSPYVSS FDSLFPSDFF DFDSTVANMP 240 EFFGNGDYGG HLFGDDCKTG ALLDGNFDFD FDLNFGYSGQ TGNDYFPDIG DIFGSDPLVA 300 L* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gcc_A | 4e-21 | 84 | 141 | 1 | 59 | ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00027 | PBM | Transfer from AT4G23750 | Download |
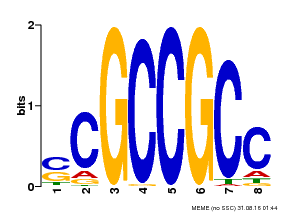
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021838632.1 | 1e-104 | ethylene-responsive transcription factor CRF2-like | ||||
| TrEMBL | A0A0K9RK75 | 1e-103 | A0A0K9RK75_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010676352.1 | 1e-95 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G23750.2 | 1e-41 | cytokinin response factor 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AHYPO_018564-RA |



