 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AHYPO_018565-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Amaranthaceae; Amaranthus
|
||||||||
| Family | YABBY | ||||||||
| Protein Properties | Length: 276aa MW: 30366.2 Da PI: 7.8438 | ||||||||
| Description | YABBY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | YABBY | 211.2 | 3.5e-65 | 33 | 232 | 3 | 170 |
YABBY 3 vfssseqvCyvqCnfCntilavsvPstslfkvvtvrCGhCtsllsvnlakas.............qllaaeshldeslkee............. 70
+ s s+q+CyvqC+fC+t+lavsvPs+slfk+vtvrCGhCt+llsv+ +l+ + +
AHYPO_018565-RA 33 LLSPSDQLCYVQCSFCDTVLAVSVPSSSLFKTVTVRCGHCTNLLSVKALLLPtatpsppyqppppTLTVNS---------Nqllhhhhhhhlsp 117
6789*****************************************99766653345566555554222223.........03333344457999 PP
YABBY 71 ........lleelkveeenlksnvekeesastsvssekl.........senedeevprvppvirPPekrqrvPsaynrfikeeiqrikasnPdi 147
llee +++ nl n++ ++++ +++++++ + +++e+p++ppv+rP ekrqrvPsaynrfik+eiqrika+nPdi
AHYPO_018565-RA 118 pfftsphnLLEEIRSSPPNLVLNNH--HQPMFNEPMNNMmplrgvdhhHHLHHQEIPKPPPVNRPQEKRQRVPSAYNRFIKDEIQRIKAGNPDI 209
9***999999999999888776654..4455555555545788887655567899*************************************** PP
YABBY 148 shreafsaaaknWahfPkihfgl 170
shreafsaaaknWahfP+ihfgl
AHYPO_018565-RA 210 SHREAFSAAAKNWAHFPHIHFGL 232
*********************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04690 | 7.4E-63 | 36 | 232 | IPR006780 | YABBY protein |
| SuperFamily | SSF47095 | 3.4E-7 | 178 | 226 | IPR009071 | High mobility group box domain |
| Gene3D | G3DSA:1.10.30.10 | 3.7E-4 | 181 | 224 | IPR009071 | High mobility group box domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009909 | Biological Process | regulation of flower development | ||||
| GO:0009933 | Biological Process | meristem structural organization | ||||
| GO:0009944 | Biological Process | polarity specification of adaxial/abaxial axis | ||||
| GO:0010093 | Biological Process | specification of floral organ identity | ||||
| GO:0010154 | Biological Process | fruit development | ||||
| GO:0010158 | Biological Process | abaxial cell fate specification | ||||
| GO:0010159 | Biological Process | specification of organ position | ||||
| GO:0010450 | Biological Process | inflorescence meristem growth | ||||
| GO:1902183 | Biological Process | regulation of shoot apical meristem development | ||||
| GO:2000024 | Biological Process | regulation of leaf development | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 276 aa Download sequence Send to blast |
MSSSSSSTTG SAGGGSGGSL DTSQIHSPSP DHLLSPSDQL CYVQCSFCDT VLAVSVPSSS 60 LFKTVTVRCG HCTNLLSVKA LLLPTATPSP PYQPPPPTLT VNSNQLLHHH HHHHLSPPFF 120 TSPHNLLEEI RSSPPNLVLN NHHQPMFNEP MNNMMPLRGV DHHHHLHHQE IPKPPPVNRP 180 QEKRQRVPSA YNRFIKDEIQ RIKAGNPDIS HREAFSAAAK NWAHFPHIHF GLMPDHQPVK 240 KANVRQQQEG DEHDVLIKEG FLAPQANVNV GVGPY* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Involved in the abaxial cell fate determination during embryogenesis and organogenesis. Regulates the initiation of embryonic shoot apical meristem (SAM) development (PubMed:10323860, PubMed:10331982, PubMed:10457020, PubMed:11812777, PubMed:12417699, PubMed:9878633, Ref.3, Ref.6, PubMed:19837869). Required during flower formation and development, particularly for the patterning of floral organs. Positive regulator of class B (AP3 and PI) activity in whorls 2 and 3. Negative regulator of class B activity in whorl 1 and of SUP activity in whorl 3. Interacts with class A proteins (AP1, AP2 and LUG) to repress class C (AG) activity in whorls 1 and 2. Contributes to the repression of KNOX genes (STM, KNAT1/BP and KNAT2) to avoid ectopic meristems. Binds DNA without sequence specificity. In vitro, can compete and displace the AP1 protein binding to DNA containing CArG box (PubMed:10323860, PubMed:10331982, PubMed:10457020, PubMed:11812777, PubMed:12417699, PubMed:9878633, Ref.3, Ref.6). {ECO:0000269|PubMed:10323860, ECO:0000269|PubMed:10331982, ECO:0000269|PubMed:10457020, ECO:0000269|PubMed:11812777, ECO:0000269|PubMed:12417699, ECO:0000269|PubMed:19837869, ECO:0000269|PubMed:9878633, ECO:0000269|Ref.3, ECO:0000269|Ref.6}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00620 | PBM | Transfer from AT2G45190 | Download |
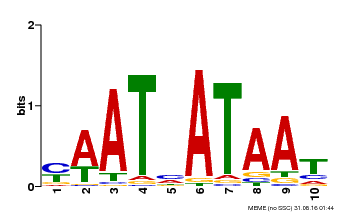
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021756313.1 | 1e-118 | axial regulator YABBY 1-like | ||||
| Swissprot | O22152 | 4e-82 | YAB1_ARATH; Axial regulator YABBY 1 | ||||
| TrEMBL | A0A0K9QN09 | 1e-103 | A0A0K9QN09_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010692186.1 | 1e-115 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G45190.1 | 2e-74 | YABBY family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AHYPO_018565-RA |



