 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AHYPO_018875-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Amaranthaceae; Amaranthus
|
||||||||
| Family | BBR-BPC | ||||||||
| Protein Properties | Length: 333aa MW: 37454.7 Da PI: 9.6545 | ||||||||
| Description | BBR-BPC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GAGA_bind | 369 | 8.5e-113 | 28 | 332 | 18 | 301 |
GAGA_bind 18 aslkenlglqlmssiaerdakirernlalsekkaavaerdmaflqrdkalaernkalverdnkllalllvensla...salpvgvqvlsgtksi 108
+s +q+ms++aerda+i+ern+a+ ek++a+ erdma +qrd+a++er++a++erdn+l+al++++n ++ +a+p+++q+++g k++
AHYPO_018875-RA 28 QS-----MKQIMSIMAERDAAIQERNMAINEKNKALEERDMAIMQRDTAIQERHEAIMERDNALAALQYRQNPMNggsNACPPNCQISRGLKHM 116
33.....35***************************************************************9999999*************** PP
GAGA_bind 109 dslqq.lse.pqledsavelreeeklealpieeaaeeakekkkkkkrqrakkpkekkakkkkkksekskkkvkkesader.............. 186
++ + +++ ++++ +++ re + +alp+ + +e+++++k+k+ ++++ +++kk++k +k+k++++d +
AHYPO_018875-RA 117 HHPEPhVQHpLHVGEASYDGREVQVPDALPLPSISSENTKSRKVKRLKEPSA------ASPNKKTSKGGRKSKRDGEDLNkimfsksqewkgme 204
**98854556899**********99999998887777777777664443332......222222222222222222221113333344578999 PP
GAGA_bind 187 .............skaekksidlvlngvslDestlPvPvCsCtGalrqCYkWGnGGWqSaCCtttiSvyPLPvstkrrgaRiagrKmSqgafkk 267
k+++k dl+ln+v++Dest+P+PvCsCtG+ rqCYkWGnGGWqS+CCtttiS+yPLP+++++r+aR++grKmS++af+k
AHYPO_018875-RA 205 dmsgsdgykqlggLKSDWKGADLGLNQVAFDESTMPPPVCSCTGVYRQCYKWGNGGWQSSCCTTTISMYPLPAVPNKRHARVGGRKMSGSAFSK 298
********************************************************************************************** PP
GAGA_bind 268 lLekLaaeGydlsnpvDLkdhWAkHGtnkfvtir 301
lL++Laa+G+dls+pvDLkdhWAkHGtn+++tir
AHYPO_018875-RA 299 LLSRLAADGHDLSTPVDLKDHWAKHGTNRYITIR 332
*********************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01226 | 3.9E-138 | 1 | 332 | IPR010409 | GAGA-binding transcriptional activator |
| Pfam | PF06217 | 1.9E-100 | 21 | 332 | IPR010409 | GAGA-binding transcriptional activator |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 333 aa Download sequence Send to blast |
MNFSTFIWWR FWILDVIFLF QWLMSQSQSM KQIMSIMAER DAAIQERNMA INEKNKALEE 60 RDMAIMQRDT AIQERHEAIM ERDNALAALQ YRQNPMNGGS NACPPNCQIS RGLKHMHHPE 120 PHVQHPLHVG EASYDGREVQ VPDALPLPSI SSENTKSRKV KRLKEPSAAS PNKKTSKGGR 180 KSKRDGEDLN KIMFSKSQEW KGMEDMSGSD GYKQLGGLKS DWKGADLGLN QVAFDESTMP 240 PPVCSCTGVY RQCYKWGNGG WQSSCCTTTI SMYPLPAVPN KRHARVGGRK MSGSAFSKLL 300 SRLAADGHDL STPVDLKDHW AKHGTNRYIT IR* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional regulator that specifically binds to GA-rich elements (GAGA-repeats) present in regulatory sequences of genes involved in developmental processes. {ECO:0000269|PubMed:14731261}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00540 | DAP | Transfer from AT5G42520 | Download |
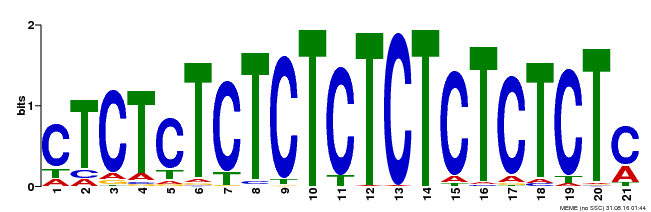
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021759836.1 | 0.0 | protein BASIC PENTACYSTEINE6-like | ||||
| Refseq | XP_021759837.1 | 0.0 | protein BASIC PENTACYSTEINE6-like | ||||
| Refseq | XP_021866753.1 | 0.0 | protein BASIC PENTACYSTEINE6 | ||||
| Refseq | XP_021866754.1 | 0.0 | protein BASIC PENTACYSTEINE6 | ||||
| Refseq | XP_021866755.1 | 0.0 | protein BASIC PENTACYSTEINE6 | ||||
| Swissprot | Q8L999 | 1e-128 | BPC6_ARATH; Protein BASIC PENTACYSTEINE6 | ||||
| TrEMBL | A0A0K9R5U0 | 0.0 | A0A0K9R5U0_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010670883.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G42520.1 | 1e-117 | basic pentacysteine 6 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AHYPO_018875-RA |



