 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AHYPO_019392-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Amaranthaceae; Amaranthus
|
||||||||
| Family | ARR-B | ||||||||
| Protein Properties | Length: 663aa MW: 73206.3 Da PI: 6.3117 | ||||||||
| Description | ARR-B family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 91.4 | 8e-29 | 209 | 262 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
kpr++W+ eLH++Fv av+qL G++kA+Pk+ilelm+v+gLt+e+v+SHLQkYRl
AHYPO_019392-RA 209 KPRVVWSVELHQQFVAAVNQL-GIDKAVPKKILELMNVPGLTRENVASHLQKYRL 262
79*******************.********************************8 PP
| |||||||
| 2 | Response_reg | 78 | 3.4e-26 | 33 | 141 | 1 | 109 |
EEEESSSHHHHHHHHHHHHHTTCEEEEEESSHHHHHHHHHHHH..ESEEEEESSCTTSEHHHHHHHHHHHTTTSEEEEEESTTTHHHHHHHHHT CS
Response_reg 1 vlivdDeplvrellrqalekegyeevaeaddgeealellkekd..pDlillDiempgmdGlellkeireeepklpiivvtahgeeedalealka 92
vl+vdD+p+ + +l+++l+ +y ev+ + +e al+ll+e++ +D+++ D+ mp+mdG++ll++ e +lp+i+++a +++ + + +
AHYPO_019392-RA 33 VLVVDDDPTCLTILERMLRTCRY-EVTKTNRAENALTLLRENRngFDVVISDVHMPDMDGFKLLEHVGLEM-DLPVIMMSADDSKHVVMKGVTH 124
89*********************.***************99999***********************6644.8********************* PP
TESEEEESS--HHHHHH CS
Response_reg 93 GakdflsKpfdpeelvk 109
Ga d+l Kp+ +e+l +
AHYPO_019392-RA 125 GACDYLIKPVRIEALKN 141
*************9986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PIRSF | PIRSF036392 | 2.8E-171 | 10 | 659 | IPR017053 | Response regulator B-type, plant |
| Gene3D | G3DSA:3.40.50.2300 | 8.9E-42 | 30 | 168 | No hit | No description |
| SuperFamily | SSF52172 | 1.57E-35 | 30 | 155 | IPR011006 | CheY-like superfamily |
| SMART | SM00448 | 3.4E-29 | 31 | 143 | IPR001789 | Signal transduction response regulator, receiver domain |
| PROSITE profile | PS50110 | 41.344 | 32 | 147 | IPR001789 | Signal transduction response regulator, receiver domain |
| Pfam | PF00072 | 3.5E-23 | 33 | 141 | IPR001789 | Signal transduction response regulator, receiver domain |
| CDD | cd00156 | 3.20E-26 | 34 | 146 | No hit | No description |
| SuperFamily | SSF46689 | 3.05E-20 | 206 | 266 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 11.048 | 206 | 265 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.1E-30 | 208 | 267 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 9.4E-25 | 209 | 262 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 9.4E-8 | 211 | 261 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009873 | Biological Process | ethylene-activated signaling pathway | ||||
| GO:0010082 | Biological Process | regulation of root meristem growth | ||||
| GO:0010119 | Biological Process | regulation of stomatal movement | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0071368 | Biological Process | cellular response to cytokinin stimulus | ||||
| GO:0080113 | Biological Process | regulation of seed growth | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 663 aa Download sequence Send to blast |
MNLGGGVVGS MTLHNSTVNR KPADDNFPVG LRVLVVDDDP TCLTILERML RTCRYEVTKT 60 NRAENALTLL RENRNGFDVV ISDVHMPDMD GFKLLEHVGL EMDLPVIMMS ADDSKHVVMK 120 GVTHGACDYL IKPVRIEALK NIWQHVVRKK RTEYKDIEQS GSWDEGDRQQ KQDDTVTSPA 180 NDGSWKNAKR KSGEDDEPEE KDDSTTLKKP RVVWSVELHQ QFVAAVNQLG IDKAVPKKIL 240 ELMNVPGLTR ENVASHLQKY RLYLRRLSGV SQHNSSFMPQ DSSFSSMSSL GGIDLQTLAA 300 TGQLSAQTLA AYTRLPPTIK PGMSMPFVDQ RNLFSFENNK LRYGEPQQAQ ISNVSKQMNL 360 LHGIPTTMEP KQLAGLNQSS QTLGNLSMQT NASISHQNNS LLMQQMVPQQ SRVQIINENV 420 NCPVSRIQSS IGQSLQSNGT GSAVLPRNAV PYDPLPQAAP VVDYSVNQIS ELPGNSFPLG 480 SNPGMTSMTS KGFSQEEVCP DIKVTRGFVG NYDMFSDLQQ KPQDWQLQNP NMSYAGSSQH 540 MQSSVNVAPS VLVNQSYIPS HKNEQNGNAI AGKQMFSTGL ENQHIRTQGI NQNINSVIVN 600 HSSRVKSENI SDVVNPGNSL FSEHYSQDDL MSALLKQQQE GIGSSDYDYD GYTLDNIPSV 660 GI* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 2e-22 | 205 | 268 | 1 | 64 | ARR10-B |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that binds specifically to the DNA sequence 5'-[AG]GATT-3'. Functions as a response regulator involved in His-to-Asp phosphorelay signal transduction system. Phosphorylation of the Asp residue in the receiver domain activates the ability of the protein to promote the transcription of target genes. Could directly activate some type-A response regulators in response to cytokinins. Involved in the expression of nuclear genes for components of mitochondrial complex I. Promotes cytokinin-mediated leaf longevity. Involved in the ethylene signaling pathway in an ETR1-dependent manner and in the cytokinin signaling pathway. {ECO:0000269|PubMed:11370868, ECO:0000269|PubMed:11574878, ECO:0000269|PubMed:15282545, ECO:0000269|PubMed:16407152}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00054 | PBM | Transfer from AT4G16110 | Download |
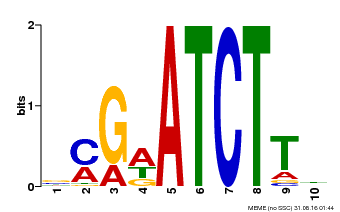
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010670914.1 | 0.0 | PREDICTED: two-component response regulator ARR2 isoform X1 | ||||
| Swissprot | Q9ZWJ9 | 0.0 | ARR2_ARATH; Two-component response regulator ARR2 | ||||
| TrEMBL | A0A0K9RPY9 | 0.0 | A0A0K9RPY9_SPIOL; Two-component response regulator | ||||
| STRING | XP_010670914.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16110.1 | 1e-176 | response regulator 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AHYPO_019392-RA |



