 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AHYPO_019672-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Amaranthaceae; Amaranthus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 717aa MW: 77702.4 Da PI: 6.7866 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 50 | 5.5e-16 | 473 | 519 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K +Ka++L +A+eY+k Lq
AHYPO_019672-RA 473 VHNLSERRRRDRINEKMRALQELIPNC-----NKADKASMLDEAIEYLKTLQ 519
6*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 4.05E-17 | 465 | 523 | No hit | No description |
| SuperFamily | SSF47459 | 1.57E-20 | 466 | 531 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 4.7E-20 | 466 | 526 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.228 | 469 | 518 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 2.4E-13 | 473 | 519 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 2.1E-17 | 475 | 524 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 717 aa Download sequence Send to blast |
MPLTEFIRML KGKPEVSQGA TTSIDPSSRP DSDCVELVWE NGQIMMQGKA RKNPISNNFQ 60 TFGSKPQDDG RVSNTSKMTK FGQMNSEMAE FPLCVPSCEM GLDQDVDLVP WLSYPMDQPL 120 HNDYTPDFLN ELSGVTINDL PSESHLAYRE RITSFHGDGG HDTSMTLKHS HTSKPPSSNV 180 KDNRGGSGEV PQFLPSFQQS VPRSGISDGN NLGNAQKAAS RDSLPNPSPL SDFASLRLHK 240 LKAGQPSTSS GITNFLEFSR PASLSRASHV MVNPSMILEN DDKRCTLSSN NPAKSALVNL 300 DSGCSQKDIV SHNQTIVEPA NANLESFPTK PNPELISTKE SLYREKLAIN QMSPNQACDT 360 SMNKAGIGGD DRIVEPVVAA SSVCSGNSVD RASNEPSRNL KRKCLETADS SSPSDDVRYW 420 ISLQTDLSLQ FCFFVWHDKL NMAYLQENED ESVGARKPVC GQTGSKKSRA AEVHNLSERR 480 RRDRINEKMR ALQELIPNCN KADKASMLDE AIEYLKTLQL QVQILSMGPG LYMQPMLFPP 540 GMQPIHGAHM QHFSPMGLGM GMGMGLGMNM LNMNSGARMV PFLGPHYPIP GIRPPFHGMP 600 GSSLQAFPHP GQGFPMSMQQ PSVPPPKAGN FMKMPSGSSA SGVAGTSNAP HFASSGCAKD 660 ANPQLMPSGV TSTNVNSSLN QATNQASNER TQQSAMSIDQ AQDVKPSEAA NKENNA* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 477 | 482 | ERRRRD |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
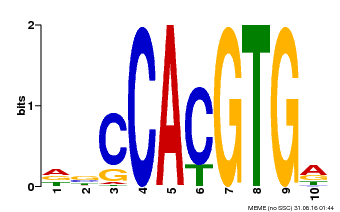
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019104506.1 | 0.0 | PREDICTED: transcription factor PIF3 isoform X1 | ||||
| Refseq | XP_019104507.1 | 0.0 | PREDICTED: transcription factor PIF3 isoform X1 | ||||
| Refseq | XP_019104508.1 | 0.0 | PREDICTED: transcription factor PIF3 isoform X1 | ||||
| TrEMBL | A0A0K9RD81 | 0.0 | A0A0K9RD81_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010674470.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 2e-42 | phytochrome interacting factor 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AHYPO_019672-RA |



