 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AHYPO_020532-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Amaranthaceae; Amaranthus
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 308aa MW: 34709.4 Da PI: 4.4403 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 24.1 | 7.8e-08 | 153 | 195 | 3 | 45 |
XXCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 3 elkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaL 45
+k+ +r +NR AA +sR+RKk +++ Le k+ +eae ++L
AHYPO_020532-RA 153 LSKKRKRQLRNRDAAMKSRERKKIYVKDLEMKSRYMEAECRRL 195
57999*********************************98766 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.20.5.170 | 2.6E-10 | 148 | 210 | No hit | No description |
| PROSITE profile | PS50217 | 9.347 | 153 | 195 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF07716 | 2.1E-7 | 154 | 195 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 7.74E-10 | 155 | 210 | No hit | No description |
| CDD | cd14704 | 6.38E-14 | 156 | 207 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 158 | 173 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0030968 | Biological Process | endoplasmic reticulum unfolded protein response | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005789 | Cellular Component | endoplasmic reticulum membrane | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 308 aa Download sequence Send to blast |
MEVEGLNEYE WEHIFHDLEI PRNGFDDVFT EASSLPATEA IDSWIEELLM NDDGEQTSAP 60 NDCFTWSDLL VDSPNENFQV SDEHPPTEVK SSPNASESEG NLVDATRNPI DYDYKSNNNG 120 DGNTRNFKSN GENDKSGDDN LEGKDIDSDA DPLSKKRKRQ LRNRDAAMKS RERKKIYVKD 180 LEMKSRYMEA ECRRLGRLLQ CCYAENQMLR ISMSNAYGAS LTKPESAVLL LESLLLVSLV 240 WLMVNMCLLS QSKVQAPHES EETKKQKQEN AAQNRRGATV TDVSPQPQPL PQSRIDLADS 300 QGTQLRV* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 155 | 174 | KRKRQLRNRDAAMKSRERKK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00019 | PBM | Transfer from AT1G42990 | Download |
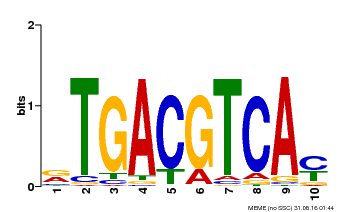
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010684737.1 | 8e-74 | PREDICTED: bZIP transcription factor 60 | ||||
| TrEMBL | A0A0K9QFM6 | 9e-64 | A0A0K9QFM6_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010684737.1 | 3e-73 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G42990.1 | 2e-13 | basic region/leucine zipper motif 60 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AHYPO_020532-RA |



