 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AHYPO_020533-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Amaranthaceae; Amaranthus
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 1101aa MW: 121963 Da PI: 7.1264 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 126.9 | 8.1e-40 | 180 | 257 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqv++C++dls+ k+yhrrhkvCe+hska+++lvs+++qrfCqqCsrfh lsefDe+krsCrrrLa+hn+rrrk+qa
AHYPO_020533-RA 180 MCQVDNCREDLSKSKDYHRRHKVCEFHSKATKALVSKQMQRFCQQCSRFHPLSEFDEGKRSCRRRLAGHNKRRRKTQA 257
6**************************************************************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 2.2E-33 | 174 | 242 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 31.65 | 178 | 255 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 9.42E-37 | 179 | 259 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 2.6E-29 | 181 | 254 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF48403 | 2.82E-8 | 851 | 990 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 9.6E-7 | 891 | 994 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 9.05E-8 | 896 | 989 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1101 aa Download sequence Send to blast |
MEEVGTQVAS PLFIHQNIGG RFCDVSLIGT KRSLCYSTTS NHQQQHQIHH PSHGWKLKDW 60 EWDSTRFVAR PRPVEPDVLC LGTLNQAPSN CNEAVNPILS TPLPSKSSNR LDDDKPAGSI 120 LRLQLGGCGI GDSDGAGTTI TKTNTGNAQN SNSKEDPVSS SRPNKKVRSG SPGSGGNYPM 180 CQVDNCREDL SKSKDYHRRH KVCEFHSKAT KALVSKQMQR FCQQCSRFHP LSEFDEGKRS 240 CRRRLAGHNK RRRKTQAEDT ASPAVQTTDN NKPGYGNLDI VNLLTVLARG QGNAELNVPT 300 CPSLPDKNQL IQILSKINSL PLPADIATNP LLSSFSARNG FEQGSLDPSN MDVNAASRST 360 TDLLAVLSST LPGSSPETLA FFSNMNNSAS SMDKNMSPIV DKDDAHKKPT VELQPLGGEK 420 SSSSYQSPTD DSDSQVQDTR LNLPLQLFSS SPGEDSSPNL ASSRKYFSSD SSNPTEERSH 480 SSTAPVTRKL FPLETTSESA RPVRMSFDEE AVVNIEQSQT ENSATRMTLE LFSMGKKTVS 540 NALLNLPPQA GYTSSSGSDH SPPSFNSDPQ KDRTGRIIFK LFDKDPSHFP GALRTQIYNW 600 LSNSPSEMES YIRPGCVVLS IYLSMKSAAW EQMEENFLQQ VEALLQSSDV EFWRSGRFSV 660 NIGRQLAIHK DGRIRICKPW STVTSPELFM VSPLAVVNGE NTTLVLRGMN LTSPGTKIHC 720 TYTGSYSSTE VLQFSDQGFT YDEIKLNEFK VYAAPSALGR CFIEVENGVR GNCFPVIIAD 780 VEICQELRLL EREFYEAKVS DVVTDVQTPY VASPHSHDEV LHFLNELGWL FQRQLCPEIA 840 VHDYMLHRFQ YLLTFSTERN YCALVKRLLD IFIESESRMD GLSTECMEAL ANMRLLHRAV 900 KGRSTKMVDL LIHYSAPCGS ITSKKYIFPP NHRGPGGITP LHLAACTSDS YEIVDVLTGD 960 PMQIGLHSWR FLLDDNGQSP YSYAVMRNDH ALNNIVGQKL ADKTNSQVSI NIGNEIVESA 1020 VPAEVRQRSS LQLNTKQNSC SKCVMMRYSR MPGSQGFLHR PFIHSMLTIA AVCVCVCLFF 1080 KSLHVNSVSF MWEHVDFGAM * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 1e-32 | 170 | 254 | 1 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' (By similarity). May play a role in plant development. {ECO:0000250, ECO:0000269|PubMed:15703061}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00155 | DAP | Transfer from AT1G20980 | Download |
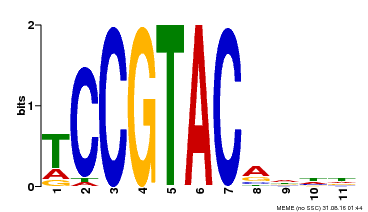
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021717801.1 | 0.0 | squamosa promoter-binding-like protein 14 | ||||
| Swissprot | Q8RY95 | 0.0 | SPL14_ARATH; Squamosa promoter-binding-like protein 14 | ||||
| TrEMBL | A0A0K9R1B9 | 0.0 | A0A0K9R1B9_SPIOL; Uncharacterized protein | ||||
| STRING | VIT_18s0122g00380.t01 | 0.0 | (Vitis vinifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G20980.1 | 0.0 | squamosa promoter binding protein-like 14 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AHYPO_020533-RA |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



