 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AT1G59750.4 | ||||||||
| Common Name | ARF1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 660aa MW: 73216.1 Da PI: 6.1534 | ||||||||
| Description | auxin response factor 1 | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 67.8 | 1.5e-21 | 124 | 225 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT.......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE. CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh.......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrsefe 91
f+k+lt sd++++g +++ +++a+++ +++ ++l+ +d+++ +W +++i+r++++r++lt+GW+ Fv++++L +gD ++F + +++e
AT1G59750.4 124 FCKTLTASDTSTHGGFSVLRRHADDClppldmsQQP--PWQELVATDLHNSEWHFRHIFRGQPRRHLLTTGWSVFVSSKKLVAGDAFIFL--RGENEE 217
99****************************964333..4459************************************************..348888 PP
.EEEEE-S CS
B3 92 lvvkvfrk 99
l+v+v+r+
AT1G59750.4 218 LRVGVRRH 225
9****996 PP
| |||||||
| 2 | Auxin_resp | 112.3 | 4.6e-37 | 250 | 332 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalkvkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsLk 83
aaha++t+++F+v+Y+Pr+s+seF+v+v+++++a ++k+svGmRfkm+fe+e+++e+r+sGt+vgv++ +++ W++S+WrsLk
AT1G59750.4 250 AAHAITTGTIFSVFYKPRTSRSEFIVSVNRYLEAKTQKLSVGMRFKMRFEGEEAPEKRFSGTIVGVQENKSSVWHDSEWRSLK 332
79*******************************************************************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101936 | 5.23E-44 | 113 | 253 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 8.4E-40 | 121 | 239 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 6.10E-19 | 122 | 224 | No hit | No description |
| PROSITE profile | PS50863 | 10.518 | 124 | 226 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 2.2E-19 | 124 | 225 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 1.6E-20 | 124 | 226 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 3.4E-34 | 250 | 332 | IPR010525 | Auxin response factor |
| Pfam | PF02309 | 3.4E-11 | 532 | 627 | IPR033389 | AUX/IAA domain |
| PROSITE profile | PS51745 | 30.767 | 537 | 630 | IPR000270 | PB1 domain |
| SuperFamily | SSF54277 | 2.62E-11 | 539 | 614 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0015031 | Biological Process | protein transport | ||||
| GO:0016192 | Biological Process | vesicle-mediated transport | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005768 | Cellular Component | endosome | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| Plant Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PO Term | PO Category | PO Description | ||||
| PO:0000013 | anatomy | cauline leaf | ||||
| PO:0000037 | anatomy | shoot apex | ||||
| PO:0000230 | anatomy | inflorescence meristem | ||||
| PO:0000293 | anatomy | guard cell | ||||
| PO:0008019 | anatomy | leaf lamina base | ||||
| PO:0009005 | anatomy | root | ||||
| PO:0009006 | anatomy | shoot system | ||||
| PO:0009009 | anatomy | plant embryo | ||||
| PO:0009010 | anatomy | seed | ||||
| PO:0009025 | anatomy | vascular leaf | ||||
| PO:0009029 | anatomy | stamen | ||||
| PO:0009030 | anatomy | carpel | ||||
| PO:0009031 | anatomy | sepal | ||||
| PO:0009032 | anatomy | petal | ||||
| PO:0009046 | anatomy | flower | ||||
| PO:0009047 | anatomy | stem | ||||
| PO:0009052 | anatomy | flower pedicel | ||||
| PO:0020030 | anatomy | cotyledon | ||||
| PO:0020038 | anatomy | petiole | ||||
| PO:0020100 | anatomy | hypocotyl | ||||
| PO:0020137 | anatomy | leaf apex | ||||
| PO:0025022 | anatomy | collective leaf structure | ||||
| PO:0001054 | developmental stage | vascular leaf senescent stage | ||||
| PO:0001078 | developmental stage | plant embryo cotyledonary stage | ||||
| PO:0001081 | developmental stage | mature plant embryo stage | ||||
| PO:0001185 | developmental stage | plant embryo globular stage | ||||
| PO:0004507 | developmental stage | plant embryo bilateral stage | ||||
| PO:0007064 | developmental stage | LP.12 twelve leaves visible stage | ||||
| PO:0007095 | developmental stage | LP.08 eight leaves visible stage | ||||
| PO:0007098 | developmental stage | LP.02 two leaves visible stage | ||||
| PO:0007103 | developmental stage | LP.10 ten leaves visible stage | ||||
| PO:0007115 | developmental stage | LP.04 four leaves visible stage | ||||
| PO:0007123 | developmental stage | LP.06 six leaves visible stage | ||||
| PO:0007611 | developmental stage | petal differentiation and expansion stage | ||||
| PO:0007616 | developmental stage | flowering stage | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 660 aa Download sequence Send to blast |
MAASNHSSGK PGGVLSDALC RELWHACAGP LVTLPREGER VYYFPEGHME QLEASMHQGL 60 EQQMPSFNLP SKILCKVINI QRRAEPETDE VYAQITLLPE LDQSEPTSPD APVQEPEKCT 120 VHSFCKTLTA SDTSTHGGFS VLRRHADDCL PPLDMSQQPP WQELVATDLH NSEWHFRHIF 180 RGQPRRHLLT TGWSVFVSSK KLVAGDAFIF LRGENEELRV GVRRHMRQQT NIPSSVISSH 240 SMHIGVLATA AHAITTGTIF SVFYKPRTSR SEFIVSVNRY LEAKTQKLSV GMRFKMRFEG 300 EEAPEKRFSG TIVGVQENKS SVWHDSEWRS LKVQWDEPSS VFRPERVSPW ELEPLVANST 360 PSSQPQPPQR NKRPRPPGLP SPATGPSDGV WKSPADTPSS VPLFSPPAKA ATFGHGGNKS 420 FGVSIGSAFW PTNADSAAES FASAFNNEST EKKQTNGNVC RLFGFELVEN VNVDECFSAA 480 SVSGAVAVDQ PVPSNEFDSG QQSEPLNINQ SDIPSGSGDP EKSSLRSPQE SQSRQIRSCT 540 KVHMQGSAVG RAIDLTRSEC YEDLFKKLEE MFDIKGELLE STKKWQVVYT DDEDDMMMVG 600 DDPWNEFCGM VRKIFIYTPE EVKKLSPKNK LAVNARMQLK ADAEENGNTE GRSSSMAGSR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldv_A | 0.0 | 1 | 359 | 1 | 359 | Auxin response factor 1 |
| 4ldw_A | 0.0 | 1 | 359 | 1 | 359 | Auxin response factor 1 |
| 4ldw_B | 0.0 | 1 | 359 | 1 | 359 | Auxin response factor 1 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 371 | 375 | KRPRP |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| At.74832 | 0.0 | bud| flower| leaf| root | ||||
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | E-value | ||||
| Genevisible | 262914_at | 0.0 | ||||
| Expression Atlas | AT1G59750 | - | ||||
| AtGenExpress | AT1G59750 | - | ||||
| ATTED-II | AT1G59750 | - | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Expressed in the sepals and carpels of young flower buds. At stage 10 of flower development, expression in the carpels becomes restricted to the style. Also expressed in anthers and filaments. At stage 13, expressed in the region at the top of the pedicel, including the abscission zone. {ECO:0000269|PubMed:16176952}. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in the whole plant. {ECO:0000269|PubMed:10476078}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| TAIR | Encodes a member of the auxin response factor family. ARFs bind to the cis element 5'-TGTCTC-3' ARFs mediate changes in gene expression in response to auxin. ARF's form heterodimers with IAA/AUX genes. ARF1 enhances mutant phenotypes of ARF2 and may act with ARF2 to control aspects of maturation and senescence.ARF1:LUC and 3xHA:ARF1 proteins have a half-life of ~3-4 hours and their degradation is reduced by proteasome inhibitors. 3xHA:ARF1 degradation is not affected by a pre-treatment with IAA. A nuclear-targeted fusion protein containing the middle region of ARF1 linked to LUC:NLS has a similar half-life to the full-length ARF1:LUC construct. The degradation of 3xHA:ARF1 is not affected in an axr6-3 mutant grown at room temperature, although the degradation of AXR2/IAA7 is slowed under these conditions. | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). Seems to act as transcriptional repressor. Formation of heterodimers with Aux/IAA proteins may alter their ability to modulate early auxin response genes expression. Promotes flowering, stamen development, floral organ abscission and fruit dehiscence. Acts as repressor of IAA2, IAA3 and IAA7. {ECO:0000269|PubMed:12036261, ECO:0000269|PubMed:16176952}. | |||||
| Function -- GeneRIF ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
|
||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00097 | PBM | 26531826 | Download |
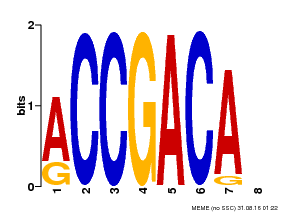
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | AT1G59750.4 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Regulation -- Hormone ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hormone | |||||
| AHD | auxin | |||||
| Interaction ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Intact With | |||||
| BioGRID | AT1G59750 | |||||
| Phenotype -- Mutation ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | |||||
| T-DNA Express | AT1G59750 | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY133723 | 0.0 | AY133723.1 Arabidopsis thaliana auxin response factor 1 (At1g59750) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001185266.1 | 0.0 | auxin response factor 1 | ||||
| Swissprot | Q8L7G0 | 0.0 | ARFA_ARATH; Auxin response factor 1 | ||||
| TrEMBL | F4ID31 | 0.0 | F4ID31_ARATH; Auxin response factor | ||||
| STRING | AT1G59750.1 | 0.0 | (Arabidopsis thaliana) | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AT1G59750.4 |
| Entrez Gene | 842268 |
| iHOP | AT1G59750 |
| wikigenes | AT1G59750 |



