 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AT1G66370.1 | ||||||||
| Common Name | AtMYB113, MYB113, T27F4.12 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 246aa MW: 28307.5 Da PI: 8.8 | ||||||||
| Description | myb domain protein 113 | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 51 | 3.4e-16 | 10 | 57 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g WTteEd ll +++ ++G g W++++ + g++R++k+c++rw++yl
AT1G66370.1 10 KGTWTTEEDILLRQCIDKYGEGKWHRVPLRTGLNRCRKSCRLRWLNYL 57
799*******************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 42.1 | 2e-13 | 69 | 108 | 7 | 48 |
HHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 7 eEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+E +l ++++k+lG++ W++Ia +++ gRt++++k++w+++l
AT1G66370.1 69 DEVDLVLRLHKLLGNR-WSLIAGRLP-GRTANDVKNYWNTHL 108
788999**********.*********.************996 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 24.913 | 5 | 61 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.1E-20 | 6 | 59 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 1.22E-26 | 7 | 104 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 7.1E-14 | 9 | 59 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.5E-15 | 10 | 57 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.42E-9 | 12 | 57 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 2.2E-20 | 60 | 111 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 18.066 | 62 | 112 | IPR017930 | Myb domain |
| SMART | SM00717 | 3.9E-11 | 62 | 110 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 7.2E-12 | 69 | 108 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 5.43E-9 | 69 | 108 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006357 | Biological Process | regulation of transcription from RNA polymerase II promoter | ||||
| GO:0007275 | Biological Process | multicellular organism development | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0030154 | Biological Process | cell differentiation | ||||
| GO:0031540 | Biological Process | regulation of anthocyanin biosynthetic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0000981 | Molecular Function | RNA polymerase II transcription factor activity, sequence-specific DNA binding | ||||
| GO:0001135 | Molecular Function | transcription factor activity, RNA polymerase II transcription factor recruiting | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Plant Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PO Term | PO Category | PO Description | ||||
| PO:0000013 | anatomy | cauline leaf | ||||
| PO:0000037 | anatomy | shoot apex | ||||
| PO:0000084 | anatomy | plant sperm cell | ||||
| PO:0000230 | anatomy | inflorescence meristem | ||||
| PO:0000293 | anatomy | guard cell | ||||
| PO:0008019 | anatomy | leaf lamina base | ||||
| PO:0009005 | anatomy | root | ||||
| PO:0009006 | anatomy | shoot system | ||||
| PO:0009009 | anatomy | plant embryo | ||||
| PO:0009010 | anatomy | seed | ||||
| PO:0009025 | anatomy | vascular leaf | ||||
| PO:0009029 | anatomy | stamen | ||||
| PO:0009030 | anatomy | carpel | ||||
| PO:0009031 | anatomy | sepal | ||||
| PO:0009032 | anatomy | petal | ||||
| PO:0009046 | anatomy | flower | ||||
| PO:0009047 | anatomy | stem | ||||
| PO:0009052 | anatomy | flower pedicel | ||||
| PO:0020030 | anatomy | cotyledon | ||||
| PO:0020038 | anatomy | petiole | ||||
| PO:0020100 | anatomy | hypocotyl | ||||
| PO:0020137 | anatomy | leaf apex | ||||
| PO:0025022 | anatomy | collective leaf structure | ||||
| PO:0025281 | anatomy | pollen | ||||
| PO:0001016 | developmental stage | L mature pollen stage | ||||
| PO:0001017 | developmental stage | M germinated pollen stage | ||||
| PO:0001054 | developmental stage | vascular leaf senescent stage | ||||
| PO:0001078 | developmental stage | plant embryo cotyledonary stage | ||||
| PO:0001081 | developmental stage | mature plant embryo stage | ||||
| PO:0001185 | developmental stage | plant embryo globular stage | ||||
| PO:0004507 | developmental stage | plant embryo bilateral stage | ||||
| PO:0007064 | developmental stage | LP.12 twelve leaves visible stage | ||||
| PO:0007095 | developmental stage | LP.08 eight leaves visible stage | ||||
| PO:0007098 | developmental stage | LP.02 two leaves visible stage | ||||
| PO:0007103 | developmental stage | LP.10 ten leaves visible stage | ||||
| PO:0007115 | developmental stage | LP.04 four leaves visible stage | ||||
| PO:0007123 | developmental stage | LP.06 six leaves visible stage | ||||
| PO:0007611 | developmental stage | petal differentiation and expansion stage | ||||
| PO:0007616 | developmental stage | flowering stage | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 246 aa Download sequence Send to blast |
MGESPKGLRK GTWTTEEDIL LRQCIDKYGE GKWHRVPLRT GLNRCRKSCR LRWLNYLKPS 60 IKRGKLCSDE VDLVLRLHKL LGNRWSLIAG RLPGRTANDV KNYWNTHLSK KHDERCCKTK 120 MINKNITSHP TSSAQKIDVL KPRPRSFSDK NSCNDVNILP KVDVVPLHLG LNNNYVCESS 180 ITCNKDEQKD KLININLLDG DNMWWESLLE ADVLGPEATE TAKGVTLPLD FEQIWARFDE 240 ETLELN |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 2e-21 | 8 | 113 | 5 | 109 | B-MYB |
| Search in ModeBase | ||||||
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | E-value | ||||
| GEO | 11641121 | 0.0 | ||||
| Genevisible | 260134_at | 0.0 | ||||
| Expression Atlas | AT1G66370 | - | ||||
| AtGenExpress | AT1G66370 | - | ||||
| ATTED-II | AT1G66370 | - | ||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| TAIR | Encodes a member of the MYB family of transcription factors. Involved in regulation of anthocyanin biosynthesis. Affects the expression of enzymes involved in later steps of anthocyanin biosynthesis. | |||||
| UniProt | Transcription activator, when associated with BHLH002/EGL3/MYC146, BHLH012/MYC1, or BHLH042/TT8. {ECO:0000269|PubMed:15361138}. | |||||
| Function -- GeneRIF ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
|
||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00213 | DAP | 27203113 | Download |
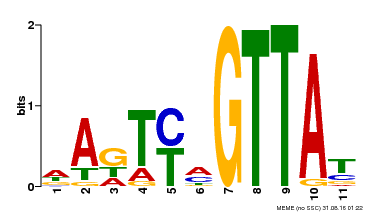
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | AT1G66370.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Regulation -- Hormone ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hormone | |||||
| AHD | jasmonic acid | |||||
| Interaction ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Intact With | |||||
| IntAct | Search Q9FNV9 | |||||
| Phenotype -- Mutation ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | |||||
| T-DNA Express | AT1G66370 | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY008378 | 0.0 | AY008378.1 Arabidopsis thaliana putative transcription factor MYB113 mRNA, complete cds. | |||
| GenBank | AY519566 | 0.0 | AY519566.1 Arabidopsis thaliana MYB transcription factor (At1g66370) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_176811.1 | 0.0 | myb domain protein 113 | ||||
| Swissprot | Q9FNV9 | 0.0 | MY113_ARATH; Transcription factor MYB113 | ||||
| TrEMBL | A0A178W1H8 | 1e-179 | A0A178W1H8_ARATH; MYB113 | ||||
| STRING | AT1G66370.1 | 1e-180 | (Arabidopsis thaliana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4 | 28 | 2646 | Representative plant | OGRP5 | 17 | 1784 |
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AT1G66370.1 |
| Entrez Gene | 842955 |
| iHOP | AT1G66370 |
| wikigenes | AT1G66370 |



