 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AT1G77640.1 | ||||||||
| Common Name | ERF013, T5M16.23 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 244aa MW: 26981.4 Da PI: 4.8302 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 57 | 4.9e-18 | 41 | 91 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
++ykGVr ++ +g+Wv eIr p ++++r++lg++ tae Aa+a++aa ++l+g
AT1G77640.1 41 KKYKGVRMRS-WGSWVTEIRAP---NQKTRIWLGSYSTAEAAARAYDAALLCLKG 91
59*****998.**********9...336*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 7.71E-18 | 41 | 100 | No hit | No description |
| SuperFamily | SSF54171 | 4.12E-19 | 42 | 99 | IPR016177 | DNA-binding domain |
| Gene3D | G3DSA:3.30.730.10 | 5.5E-30 | 42 | 100 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 3.0E-34 | 42 | 105 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 21.812 | 42 | 99 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 1.2E-11 | 42 | 91 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 5.9E-9 | 43 | 54 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 5.9E-9 | 65 | 81 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009873 | Biological Process | ethylene-activated signaling pathway | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Plant Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PO Term | PO Category | PO Description | ||||
| PO:0000037 | anatomy | shoot apex | ||||
| PO:0000230 | anatomy | inflorescence meristem | ||||
| PO:0009005 | anatomy | root | ||||
| PO:0009025 | anatomy | vascular leaf | ||||
| PO:0009030 | anatomy | carpel | ||||
| PO:0009046 | anatomy | flower | ||||
| PO:0009047 | anatomy | stem | ||||
| PO:0020030 | anatomy | cotyledon | ||||
| PO:0020038 | anatomy | petiole | ||||
| PO:0020100 | anatomy | hypocotyl | ||||
| PO:0020137 | anatomy | leaf apex | ||||
| PO:0025022 | anatomy | collective leaf structure | ||||
| PO:0007064 | developmental stage | LP.12 twelve leaves visible stage | ||||
| PO:0007095 | developmental stage | LP.08 eight leaves visible stage | ||||
| PO:0007098 | developmental stage | LP.02 two leaves visible stage | ||||
| PO:0007103 | developmental stage | LP.10 ten leaves visible stage | ||||
| PO:0007115 | developmental stage | LP.04 four leaves visible stage | ||||
| PO:0007123 | developmental stage | LP.06 six leaves visible stage | ||||
| PO:0007611 | developmental stage | petal differentiation and expansion stage | ||||
| PO:0007616 | developmental stage | flowering stage | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 244 aa Download sequence Send to blast |
MVKQELKIQV TTSSSSLSHS SSSSSSSTSA LRHQSCKNKI KKYKGVRMRS WGSWVTEIRA 60 PNQKTRIWLG SYSTAEAAAR AYDAALLCLK GPKANLNFPN ITTTSPFLMN IDEKTLLSPK 120 SIQKVAAQAA NSSSDHFTPP SDENDHDHDD GLDHHPSASS SAASSPPDDD HHNDDDGDLV 180 SLMESFVDYN EHVSLMDPSL YEFGHNEIFF TNGDPFDYSP QLHSSEATMD DFYDDVDIPL 240 WSFS |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| At.17793 | 0.0 | flower| root | ||||
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | E-value | ||||
| GEO | 145337680 | 0.0 | ||||
| Genevisible | 259729_at | 0.0 | ||||
| Expression Atlas | AT1G77640 | - | ||||
| AtGenExpress | AT1G77640 | - | ||||
| ATTED-II | AT1G77640 | - | ||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| TAIR | encodes a member of the DREB subfamily A-5 of ERF/AP2 transcription factor family. The protein contains one AP2 domain. There are 15 members in this subfamily including RAP2.1, RAP2.9 and RAP2.10. | |||||
| UniProt | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00246 | DAP | 27203113 | Download |
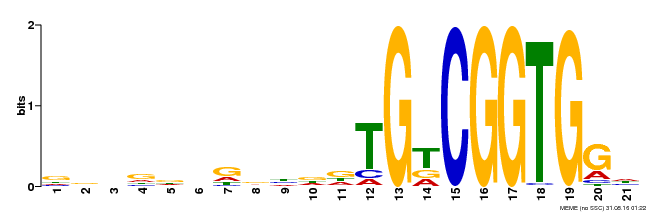
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | AT1G77640.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Phenotype -- Mutation ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | |||||
| T-DNA Express | AT1G77640 | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC010704 | 0.0 | AC010704.6 Arabidopsis thaliana chromosome 1 BAC T5M16 genomic sequence, complete sequence. | |||
| GenBank | AY560843 | 0.0 | AY560843.1 Arabidopsis thaliana putative AP2/EREBP transcription factor (At1g77640) mRNA, complete cds. | |||
| GenBank | BT012629 | 0.0 | BT012629.1 Arabidopsis thaliana At1g77640 mRNA, complete cds. | |||
| GenBank | CP002684 | 0.0 | CP002684.1 Arabidopsis thaliana chromosome 1 sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_177887.1 | 1e-180 | Integrase-type DNA-binding superfamily protein | ||||
| Swissprot | Q9CAP4 | 0.0 | ERF13_ARATH; Ethylene-responsive transcription factor ERF013 | ||||
| TrEMBL | A0A178WBA4 | 1e-177 | A0A178WBA4_ARATH; Uncharacterized protein | ||||
| STRING | AT1G77640.1 | 1e-180 | (Arabidopsis thaliana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2999 | 27 | 67 | Representative plant | OGRP6 | 16 | 1718 |
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AT1G77640.1 |
| Entrez Gene | 844099 |
| iHOP | AT1G77640 |
| wikigenes | AT1G77640 |



