 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AT4G01550.1 | ||||||||
| Common Name | ANAC069, F11O4.5, NAC069, NAC69, NTL13, NTM2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 457aa MW: 51816 Da PI: 5.26 | ||||||||
| Description | NAC domain containing protein 69 | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 136.4 | 1.8e-42 | 6 | 135 | 3 | 128 |
NAM 3 pGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLp..kkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevlsk..k 96
+G+rF Pt eel+++yLk+k+ gk+ ++e+i+e++i++++P Lp +k+k+++ wyfF++++ ++a++k ++r+t+sgyWkatg d+++++k +
AT4G01550.1 6 VGYRFYPTGEELINHYLKNKILGKTWLVDEAISEINICSYDPIYLPslSKIKSDDPVWYFFCPKEYTSAKKKVTKRTTSSGYWKATGVDRKIKDKrgN 103
7************************999999***************5449999999**************************************9666 PP
NAM 97 gelvglkktLvfykgrapkgektdWvmheyrl 128
+ ++g+kktLv+y+gr pkg+ t Wvmhey++
AT4G01550.1 104 RGEIGIKKTLVYYEGRVPKGVWTPWVMHEYHI 135
667***************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51005 | 45.577 | 4 | 153 | IPR003441 | NAC domain |
| SuperFamily | SSF101941 | 1.83E-46 | 5 | 153 | IPR003441 | NAC domain |
| Pfam | PF02365 | 7.5E-25 | 6 | 135 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0007275 | Biological Process | multicellular organism development | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0016021 | Cellular Component | integral component of membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Plant Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PO Term | PO Category | PO Description | ||||
| PO:0000013 | anatomy | cauline leaf | ||||
| PO:0000037 | anatomy | shoot apex | ||||
| PO:0000084 | anatomy | plant sperm cell | ||||
| PO:0000230 | anatomy | inflorescence meristem | ||||
| PO:0000293 | anatomy | guard cell | ||||
| PO:0008019 | anatomy | leaf lamina base | ||||
| PO:0009005 | anatomy | root | ||||
| PO:0009006 | anatomy | shoot system | ||||
| PO:0009009 | anatomy | plant embryo | ||||
| PO:0009010 | anatomy | seed | ||||
| PO:0009025 | anatomy | vascular leaf | ||||
| PO:0009029 | anatomy | stamen | ||||
| PO:0009030 | anatomy | carpel | ||||
| PO:0009031 | anatomy | sepal | ||||
| PO:0009032 | anatomy | petal | ||||
| PO:0009046 | anatomy | flower | ||||
| PO:0009047 | anatomy | stem | ||||
| PO:0009052 | anatomy | flower pedicel | ||||
| PO:0020030 | anatomy | cotyledon | ||||
| PO:0020038 | anatomy | petiole | ||||
| PO:0020100 | anatomy | hypocotyl | ||||
| PO:0020137 | anatomy | leaf apex | ||||
| PO:0025022 | anatomy | collective leaf structure | ||||
| PO:0025281 | anatomy | pollen | ||||
| PO:0001054 | developmental stage | vascular leaf senescent stage | ||||
| PO:0001078 | developmental stage | plant embryo cotyledonary stage | ||||
| PO:0001081 | developmental stage | mature plant embryo stage | ||||
| PO:0001185 | developmental stage | plant embryo globular stage | ||||
| PO:0004507 | developmental stage | plant embryo bilateral stage | ||||
| PO:0007064 | developmental stage | LP.12 twelve leaves visible stage | ||||
| PO:0007095 | developmental stage | LP.08 eight leaves visible stage | ||||
| PO:0007098 | developmental stage | LP.02 two leaves visible stage | ||||
| PO:0007103 | developmental stage | LP.10 ten leaves visible stage | ||||
| PO:0007115 | developmental stage | LP.04 four leaves visible stage | ||||
| PO:0007123 | developmental stage | LP.06 six leaves visible stage | ||||
| PO:0007611 | developmental stage | petal differentiation and expansion stage | ||||
| PO:0007616 | developmental stage | flowering stage | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 457 aa Download sequence Send to blast |
MVKDLVGYRF YPTGEELINH YLKNKILGKT WLVDEAISEI NICSYDPIYL PSLSKIKSDD 60 PVWYFFCPKE YTSAKKKVTK RTTSSGYWKA TGVDRKIKDK RGNRGEIGIK KTLVYYEGRV 120 PKGVWTPWVM HEYHITCLPQ DQRNYVICQV MYKGEDGDVP SGGNNSSEPS QSLVSDSNTV 180 RATSPTALEF EKPGQENFFG MSVDDLGTPK NEQEDFSLWD VLDPDMLFSD NNNPTVHPQA 240 PHLTPNDDEF LGGLRHVNRE QVEYLFANED FISRPTLSMT ENRNDHRPKK ALSGIIVDYS 300 SDSNSDAESI SATSYQGTSS PGDDSVGSSN RQFLQTGGDE ILSSCNDLQT YGEPSISSST 360 RQSQLTRSII RPKQEVKQDT SRAVDSDTSI DKESSMVKTE KKSWFITEEA MERNRNNPRY 420 IYLMRMIIGF ILLLALISNI ISVLQNLNPA MKFDRER |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 8e-25 | 7 | 153 | 20 | 165 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 8e-25 | 7 | 153 | 20 | 165 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 8e-25 | 7 | 153 | 20 | 165 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 8e-25 | 7 | 153 | 20 | 165 | NO APICAL MERISTEM PROTEIN |
| 4dul_A | 8e-25 | 7 | 153 | 20 | 165 | NAC domain-containing protein 19 |
| 4dul_B | 8e-25 | 7 | 153 | 20 | 165 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| At.34408 | 0.0 | flower| seed | ||||
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | E-value | ||||
| Genevisible | 255585_at | 0.0 | ||||
| Expression Atlas | AT4G01550 | - | ||||
| AtGenExpress | AT4G01550 | - | ||||
| ATTED-II | AT4G01550 | - | ||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator activated by proteolytic cleavage through regulated intramembrane proteolysis (RIP). Involved in salt stress response during seed germination and seedling growth. Binds the auxin-responsive IAA30 gene promoter and may serve as a molecular link that interconnects a developmental feedback loop of auxin signaling with a salt signal transduction pathway during seed germination (PubMed:21450938). {ECO:0000269|PubMed:21450938}. | |||||
| Function -- GeneRIF ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
|
||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00426 | DAP | 27203113 | Download |
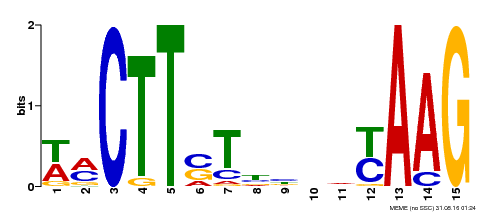
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | AT4G01550.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salt stress and abscisic acid (ABA) in roots. {ECO:0000269|PubMed:21450938}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Phenotype -- Disruption Phenotype ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | DISRUPTION PHENOTYPE: No visible phenotype under normal growth conditions, but mutant plants mutant exhibit reduced sensitivity to high salinity during seed germination and seedling development. {ECO:0000269|PubMed:21450938}. | |||||
| Phenotype -- Mutation ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | |||||
| T-DNA Express | AT4G01550 | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK229312 | 0.0 | AK229312.1 Arabidopsis thaliana mRNA for putative NAM-like protein, complete cds, clone: RAFL16-57-J19. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_192064.1 | 0.0 | NAC domain containing protein 69 | ||||
| Swissprot | Q9M126 | 0.0 | NAC69_ARATH; NAC domain-containing protein 69 | ||||
| TrEMBL | A0A178V081 | 0.0 | A0A178V081_ARATH; NTM2 | ||||
| STRING | AT4G01550.1 | 0.0 | (Arabidopsis thaliana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1982 | 19 | 71 |
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AT4G01550.1 |
| Entrez Gene | 828144 |
| iHOP | AT4G01550 |
| wikigenes | AT4G01550 |



