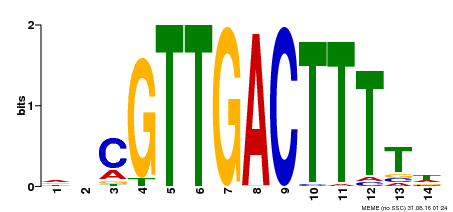
| Signature Domain? help Back to Top |
 |
| No. |
Domain |
Score |
E-value |
Start |
End |
HMM Start |
HMM End |
| 1 | WRKY | 96.4 | 1.9e-30 | 281 | 339 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-ST.T---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsa.gCpvkkkversaedpkvveitYegeHnhe 59
+D+++WrKYGqK++kgs++pr+YY+C+s gCp++k+ver+ +d ++++tYeg+Hnh
AT4G24240.1 281 SDEFSWRKYGQKPIKGSPHPRGYYKCSSVrGCPARKHVERALDDAMMLIVTYEGDHNHA 339
69*************************988****************************6 PP
|
| Publications
? help Back to Top |
- Eulgem T,Rushton PJ,Robatzek S,Somssich IE
The WRKY superfamily of plant transcription factors.
Trends Plant Sci., 2000. 5(5): p. 199-206
[PMID:10785665] - Riechmann JL, et al.
Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes.
Science, 2000. 290(5499): p. 2105-10
[PMID:11118137] - Yu D,Chen C,Chen Z
Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression.
Plant Cell, 2001. 13(7): p. 1527-40
[PMID:11449049] - Robatzek S,Somssich IE
A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes.
Plant J., 2001. 28(2): p. 123-33
[PMID:11722756] - Yamada K, et al.
Empirical analysis of transcriptional activity in the Arabidopsis genome.
Science, 2003. 302(5646): p. 842-6
[PMID:14593172] - Hoth S, et al.
Monitoring genome-wide changes in gene expression in response to endogenous cytokinin reveals targets in Arabidopsis thaliana.
FEBS Lett., 2003. 554(3): p. 373-80
[PMID:14623097] - Navarro L, et al.
The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis.
Plant Physiol., 2004. 135(2): p. 1113-28
[PMID:15181213] - Sottosanto JB,Gelli A,Blumwald E
DNA array analyses of Arabidopsis thaliana lacking a vacuolar Na+/H+ antiporter: impact of AtNHX1 on gene expression.
Plant J., 2004. 40(5): p. 752-71
[PMID:15546358] - Park CY, et al.
WRKY group IId transcription factors interact with calmodulin.
FEBS Lett., 2005. 579(6): p. 1545-50
[PMID:15733871] - Phelps-Durr TL,Thomas J,Vahab P,Timmermans MC
Maize rough sheath2 and its Arabidopsis orthologue ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis.
Plant Cell, 2005. 17(11): p. 2886-98
[PMID:16243907] - Truman W,de Zabala MT,Grant M
Type III effectors orchestrate a complex interplay between transcriptional networks to modify basal defence responses during pathogenesis and resistance.
Plant J., 2006. 46(1): p. 14-33
[PMID:16553893] - Thilmony R,Underwood W,He SY
Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157:H7.
Plant J., 2006. 46(1): p. 34-53
[PMID:16553894] - Michel K,Abderhalden O,Bruggmann R,Dudler R
Transcriptional changes in powdery mildew infected wheat and Arabidopsis leaves undergoing syringolin-triggered hypersensitive cell death at infection sites.
Plant Mol. Biol., 2006. 62(4-5): p. 561-78
[PMID:16941219] - Kim KC,Fan B,Chen Z
Pathogen-induced Arabidopsis WRKY7 is a transcriptional repressor and enhances plant susceptibility to Pseudomonas syringae.
Plant Physiol., 2006. 142(3): p. 1180-92
[PMID:16963526] - Journot-Catalino N,Somssich IE,Roby D,Kroj T
The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana.
Plant Cell, 2006. 18(11): p. 3289-302
[PMID:17114354] - Qutob D, et al.
Phytotoxicity and innate immune responses induced by Nep1-like proteins.
Plant Cell, 2006. 18(12): p. 3721-44
[PMID:17194768] - Chawade A,Br
Putative cold acclimation pathways in Arabidopsis thaliana identified by a combined analysis of mRNA co-expression patterns, promoter motifs and transcription factors.
BMC Genomics, 2007. 8: p. 304
[PMID:17764576] - Arabidopsis Interactome Mapping Consortium
Evidence for network evolution in an Arabidopsis interactome map.
Science, 2011. 333(6042): p. 601-7
[PMID:21798944] - Jin J, et al.
An Arabidopsis Transcriptional Regulatory Map Reveals Distinct Functional and Evolutionary Features of Novel Transcription Factors.
Mol. Biol. Evol., 2015. 32(7): p. 1767-73
[PMID:25750178]
|