 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AT4G38910.1 | ||||||||
| Common Name | ATBPC5, BBR/BPC5, BPC5, F19H22.10 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | BBR-BPC | ||||||||
| Protein Properties | Length: 258aa MW: 28922.9 Da PI: 10.0478 | ||||||||
| Description | basic pentacysteine 5 | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GAGA_bind | 284.1 | 6.2e-87 | 6 | 258 | 16 | 301 |
GAGA_bind 16 paaslkenlglqlmssiaerdakirernlalsekkaavaerdmaflqrdkalaernkalverdnkllalllvenslasalpvgvqvlsgtksidslqq 113
++++l ++++++s++aerda+++ern a++++k+a+a+rd+a+ qrdkal+er++a++e +++l al ++en+l+ l s +k s q
AT4G38910.1 6 QHNALV--MNKKIISILAERDAAVKERNEAVAATKEALASRDEALEQRDKALSERDNAIMETESALNALRYRENNLNYIL-------SCAKRGGS-Q- 92
456666..99****************************************************************955433.......22222221.1. PP
GAGA_bind 114 lsepqledsavelreeeklealpieeaaeeakekkkkkkrqrakkpkekkakkkkkksekskkkvkkesaderskaekksidlvlngvslDestlPvP 211
+ ++e++ + pi++ ++ea +++ +k ++ke+k+ k k +e+ ++ v++++ ++s+++++s d+ v++De t+PvP
AT4G38910.1 93 ---------RFITEESHLPNPSPISTIPPEAANTRPTK------RKKESKQGK--KMGEDLNRPVASPG--KKSRKDWDSNDV---LVTFDEMTMPVP 168
.........22344566667889999999999888777......445555544..44578999999999..569********9...589********* PP
GAGA_bind 212 vCsCtGalrqCYkWGnGGWqSaCCtttiSvyPLPvstkrrgaRiagrKmSqgafkklLekLaaeGydlsnpvDLkdhWAkHGtnkfvtir 301
+C+CtG+ rqCYkWGnGGWqS+CCttt+S yPLP+++++r++R++grKmS+++f++lL++La+eG++ls pvDLk++WA+HGtn+++ti+
AT4G38910.1 169 MCTCTGTARQCYKWGNGGWQSSCCTTTLSEYPLPQMPNKRHSRVGGRKMSGSVFSRLLSRLAGEGHELSSPVDLKNYWARHGTNRYITIK 258
*****************************************************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01226 | 2.1E-124 | 1 | 258 | IPR010409 | GAGA-binding transcriptional activator |
| Pfam | PF06217 | 2.0E-84 | 6 | 258 | IPR010409 | GAGA-binding transcriptional activator |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Plant Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PO Term | PO Category | PO Description | ||||
| PO:0000293 | anatomy | guard cell | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 258 aa Download sequence Send to blast |
MPKKEQHNAL VMNKKIISIL AERDAAVKER NEAVAATKEA LASRDEALEQ RDKALSERDN 60 AIMETESALN ALRYRENNLN YILSCAKRGG SQRFITEESH LPNPSPISTI PPEAANTRPT 120 KRKKESKQGK KMGEDLNRPV ASPGKKSRKD WDSNDVLVTF DEMTMPVPMC TCTGTARQCY 180 KWGNGGWQSS CCTTTLSEYP LPQMPNKRHS RVGGRKMSGS VFSRLLSRLA GEGHELSSPV 240 DLKNYWARHG TNRYITIK |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| At.49413 | 0.0 | flower | ||||
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | E-value | ||||
| GEO | 240256232 | 0.0 | ||||
| Expression Atlas | AT4G38910 | - | ||||
| AtGenExpress | AT4G38910 | - | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in seedlings, leaves and pistils. {ECO:0000269|PubMed:14731261}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| TAIR | The current TAIR8 structure of this gene is incorrect. The corrected structure, which will be visible in TAIR9, indicates that this gene could possibly be a pseudogene. | |||||
| UniProt | Transcriptional regulator that specifically binds to GA-rich elements (GAGA-repeats) present in regulatory sequences of genes involved in developmental processes. {ECO:0000269|PubMed:14731261}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00480 | DAP | 27203113 | Download |
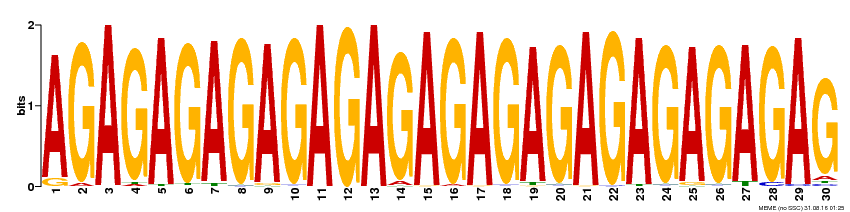
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | AT4G38910.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Phenotype -- Mutation ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | |||||
| T-DNA Express | AT4G38910 | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AL035679 | 0.0 | AL035679.1 Arabidopsis thaliana DNA chromosome 4, BAC clone F19H22 (ESSA project). | |||
| GenBank | AL161594 | 0.0 | AL161594.2 Arabidopsis thaliana DNA chromosome 4, contig fragment No. 90. | |||
| GenBank | CP002687 | 0.0 | CP002687.1 Arabidopsis thaliana chromosome 4 sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001190957.1 | 0.0 | basic pentacysteine 5 | ||||
| Refseq | NP_195602.5 | 0.0 | basic pentacysteine 5 | ||||
| Swissprot | F4JUI3 | 0.0 | BPC5_ARATH; Protein BASIC PENTACYSTEINE5 | ||||
| TrEMBL | A0A178UVL3 | 0.0 | A0A178UVL3_ARATH; BPC5 | ||||
| STRING | AT4G38910.2 | 0.0 | (Arabidopsis thaliana) | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AT4G38910.1 |
| Entrez Gene | 830046 |
| iHOP | AT4G38910 |
| wikigenes | AT4G38910 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



