 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AT5G02460.1 | ||||||||
| Common Name | DOF5.1, T1E22.3, T22P11_50 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | Dof | ||||||||
| Protein Properties | Length: 399aa MW: 42530.8 Da PI: 9.177 | ||||||||
| Description | Dof family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-Dof | 121.3 | 3.5e-38 | 92 | 151 | 3 | 62 |
zf-Dof 3 ekalkcprCdstntkfCyynnyslsqPryfCkaCrryWtkGGalrnvPvGggrrknkkss 62
e+alkcprCdstntkfCy+nnysl+qPr+fCkaCrryWt+GGalr vPvGgg+r+nk+++
AT5G02460.1 92 ETALKCPRCDSTNTKFCYFNNYSLTQPRHFCKACRRYWTRGGALRSVPVGGGCRRNKRTK 151
6789*****************************************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| ProDom | PD007478 | 4.0E-39 | 75 | 149 | IPR003851 | Zinc finger, Dof-type |
| Pfam | PF02701 | 4.0E-32 | 94 | 149 | IPR003851 | Zinc finger, Dof-type |
| PROSITE profile | PS50884 | 29.394 | 95 | 149 | IPR003851 | Zinc finger, Dof-type |
| PROSITE pattern | PS01361 | 0 | 97 | 133 | IPR003851 | Zinc finger, Dof-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Plant Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PO Term | PO Category | PO Description | ||||
| PO:0000293 | anatomy | guard cell | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 399 aa Download sequence Send to blast |
MVFSSFPTYP DHSSNWQQQH QPITTTVGFT GNNINQQFLP HHPLPPQQQQ TPPQLHHNNG 60 NGGVAVPGGP GGLIRPGSMA ERARLANIPL PETALKCPRC DSTNTKFCYF NNYSLTQPRH 120 FCKACRRYWT RGGALRSVPV GGGCRRNKRT KNSSGGGGGS TSSGNSKSQD SATSNDQYHH 180 RAMANNQMGP PSSSSSLSSL LSSYNAGLIP GHDHNSNNNN ILGLGSSLPP LKLMPPLDFT 240 DNFTLQYGAV SAPSYHIGGG SSGGAAALLN GFDQWRFPAT NQLPLGGLDP FDQQHQMEQQ 300 NPGYGLVTGS GQYRPKNIFH NLISSSSSAS SAMVTATASQ LASVKMEDSN NQLNLSRQLF 360 GDEQQLWNIH GAAAASTAAA TSSWSEVSNN FSSSSTSNI |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| At.33412 | 0.0 | flower| silique | ||||
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | E-value | ||||
| GEO | 145357549 | 0.0 | ||||
| Genevisible | 251051_at | 0.0 | ||||
| Expression Atlas | AT5G02460 | - | ||||
| AtGenExpress | AT5G02460 | - | ||||
| ATTED-II | AT5G02460 | - | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Specific to both elongating procambial cells and to cells isodiametric in size that are presumably destined to develop into higher-order veins. Strongly expressed in advance of perceptible procambium formation. Expressed in the cells of prospective veins in leaf primordia of seedlings and cotyledons of developing embryos, and the vascular tissue of developing flower buds. {ECO:0000269|PubMed:17583520}. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed ubiquitously, especially in the vascular tissues, except in seeds, petals and anthers (PubMed:20807212). Specific to the vascular tissues in young leaves, cotyledons and flower buds (PubMed:17583520). The PEAR proteins (e.g. DOF2.4, DOF5.1, DOF3.2, DOF1.1, DOF5.6 and DOF5.3) form a short-range concentration gradient that peaks at protophloem sieve elements (PSE) (PubMed:30626969). {ECO:0000269|PubMed:17583520, ECO:0000269|PubMed:20807212, ECO:0000269|PubMed:30626969}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
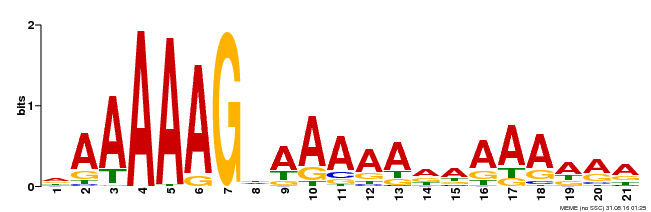
| UniProt | Transcription factor that binds specifically to a 5'-AA[AG]G-3' consensus core sequence (By similarity). Binds to 5'-TAAAGT-3' motif in REV promoter to triggers its transcription, thus regulating adaxial-abaxial polarity and influencing leaf axial patterning in an auxin transport- and response-dependent manner (e.g. IAA6 and IAA19 genes expression) (PubMed:20807212). Probably involved in early processes for vascular development (PubMed:17583520). The PEAR proteins (e.g. DOF2.4, DOF5.1, DOF3.2, DOF1.1, DOF5.6 and DOF5.3) activate gene expression that promotes radial growth of protophloem sieve elements (PubMed:30626969). {ECO:0000250|UniProtKB:Q9M2U1, ECO:0000269|PubMed:17583520, ECO:0000269|PubMed:20807212, ECO:0000269|PubMed:30626969}. | |||||
| Function -- GeneRIF ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
|
||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00483 | DAP | 27203113 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | AT5G02460.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By cytokinin in procambium. {ECO:0000269|PubMed:30626969}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Phenotype -- Mutation ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | |||||
| T-DNA Express | AT5G02460 | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AB493731 | 0.0 | AB493731.1 Arabidopsis thaliana At5g02460 mRNA for hypothetical protein, partial cds, clone: RAAt5g02460. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_195866.1 | 0.0 | Dof-type zinc finger DNA-binding family protein | ||||
| Swissprot | Q9LZ56 | 0.0 | DOF51_ARATH; Dof zinc finger protein DOF5.1 | ||||
| TrEMBL | Q6ICY3 | 0.0 | Q6ICY3_ARATH; At5g02460 | ||||
| STRING | AT5G02460.1 | 0.0 | (Arabidopsis thaliana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM11519 | 17 | 32 | Representative plant | OGRP38 | 17 | 445 |
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AT5G02460.1 |
| Entrez Gene | 830976 |
| iHOP | AT5G02460 |
| wikigenes | AT5G02460 |



