 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AT5G65410.1 | ||||||||
| Common Name | ATHB25, HB25, MNA5.14, ZFHD2, ZHD1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | ZF-HD | ||||||||
| Protein Properties | Length: 279aa MW: 31098.5 Da PI: 6.3597 | ||||||||
| Description | homeobox protein 25 | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | ZF-HD_dimer | 100.9 | 9.1e-32 | 72 | 125 | 3 | 57 |
ZF-HD_dimer 3 kvrYkeClkNhAaslGghavDGCgEfmpsegeegtaaalkCaACgCHRnFHRrev 57
++r++eClkN+A+++GghavDGCgEfmp+ g egt++alkCaACgCHRnFHR+e
AT5G65410.1 72 RFRFRECLKNQAVNIGGHAVDGCGEFMPA-GIEGTIDALKCAACGCHRNFHRKEL 125
789*************************9.999********************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04770 | 2.3E-28 | 73 | 124 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| TIGRFAMs | TIGR01566 | 6.1E-29 | 74 | 124 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| ProDom | PD125774 | 1.0E-24 | 74 | 125 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| PROSITE profile | PS51523 | 25.415 | 75 | 124 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| PROSITE pattern | PS00028 | 0 | 111 | 135 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:1.10.10.60 | 1.3E-30 | 181 | 252 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 1.85E-18 | 182 | 252 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01565 | 1.1E-25 | 191 | 247 | IPR006455 | Homeodomain, ZF-HD class |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010371 | Biological Process | regulation of gibberellin biosynthetic process | ||||
| GO:0010431 | Biological Process | seed maturation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Plant Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PO Term | PO Category | PO Description | ||||
| PO:0000013 | anatomy | cauline leaf | ||||
| PO:0000014 | anatomy | rosette leaf | ||||
| PO:0000037 | anatomy | shoot apex | ||||
| PO:0000293 | anatomy | guard cell | ||||
| PO:0008019 | anatomy | leaf lamina base | ||||
| PO:0009006 | anatomy | shoot system | ||||
| PO:0009009 | anatomy | plant embryo | ||||
| PO:0009010 | anatomy | seed | ||||
| PO:0009025 | anatomy | vascular leaf | ||||
| PO:0009031 | anatomy | sepal | ||||
| PO:0009046 | anatomy | flower | ||||
| PO:0009047 | anatomy | stem | ||||
| PO:0020038 | anatomy | petiole | ||||
| PO:0020100 | anatomy | hypocotyl | ||||
| PO:0025022 | anatomy | collective leaf structure | ||||
| PO:0001054 | developmental stage | vascular leaf senescent stage | ||||
| PO:0001078 | developmental stage | plant embryo cotyledonary stage | ||||
| PO:0001081 | developmental stage | mature plant embryo stage | ||||
| PO:0004507 | developmental stage | plant embryo bilateral stage | ||||
| PO:0007064 | developmental stage | LP.12 twelve leaves visible stage | ||||
| PO:0007098 | developmental stage | LP.02 two leaves visible stage | ||||
| PO:0007103 | developmental stage | LP.10 ten leaves visible stage | ||||
| PO:0007115 | developmental stage | LP.04 four leaves visible stage | ||||
| PO:0007611 | developmental stage | petal differentiation and expansion stage | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 279 aa Download sequence Send to blast |
MEFEDNNNNN DEEQEEDMNL HEEEEDDDAV YDSPPLSRVL PKASTESHET TGTTSTGGGG 60 GFMVVHGGGG SRFRFRECLK NQAVNIGGHA VDGCGEFMPA GIEGTIDALK CAACGCHRNF 120 HRKELPYFHH APPQHQPPPP PPGFYRLPAP VSYRPPPSQA PPLQLALPPP QRERSEDPME 180 TSSAEAGGGI RKRHRTKFTA EQKERMLALA ERIGWRIQRQ DDEVIQRFCQ ETGVPRQVLK 240 VWLHNNKHTL GKSPSPLHHH QAPPPPPPQS SFHHEQDQP |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wh5_A | 1e-43 | 182 | 249 | 8 | 75 | ZF-HD homeobox family protein |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| At.43744 | 0.0 | seed | ||||
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | E-value | ||||
| GEO | 30698120 | 0.0 | ||||
| Genevisible | 247182_at | 0.0 | ||||
| Expression Atlas | AT5G65410 | - | ||||
| AtGenExpress | AT5G65410 | - | ||||
| ATTED-II | AT5G65410 | - | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Mostly expressed in flowers and inflorescence. {ECO:0000269|PubMed:16428600}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| TAIR | Encodes ZFHD2, a member of the zinc finger homeodomain transcriptional factor family. | |||||
| UniProt | Putative transcription factor. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00588 | DAP | 27203113 | Download |
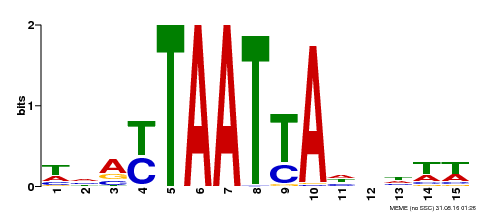
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | AT5G65410.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Interaction ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Intact With | |||||
| BioGRID | AT1G14440, AT1G26960, AT1G69600, AT1G75240 | |||||
| IntAct | Search Q9FKP8 | |||||
| Phenotype -- Mutation ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | |||||
| T-DNA Express | AT5G65410 | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AB011479 | 0.0 | AB011479.1 Arabidopsis thaliana genomic DNA, chromosome 5, P1 clone:MNA5. | |||
| GenBank | BT002385 | 0.0 | BT002385.1 Arabidopsis thaliana putative protein (At5g65410) mRNA, complete cds. | |||
| GenBank | BT006304 | 0.0 | BT006304.1 Arabidopsis thaliana At5g65410 mRNA, complete cds. | |||
| GenBank | CP002688 | 0.0 | CP002688.1 Arabidopsis thaliana chromosome 5 sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_201344.1 | 0.0 | homeobox protein 25 | ||||
| Swissprot | Q9FKP8 | 0.0 | ZHD1_ARATH; Zinc-finger homeodomain protein 1 | ||||
| TrEMBL | A0A178U7K0 | 1e-152 | A0A178U7K0_ARATH; ZHD1 | ||||
| STRING | AT5G65410.1 | 0.0 | (Arabidopsis thaliana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM13237 | 17 | 28 | Representative plant | OGRP91 | 16 | 237 |
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AT5G65410.1 |
| Entrez Gene | 836666 |
| iHOP | AT5G65410 |
| wikigenes | AT5G65410 |



