 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Achn049821 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; Ericales; Actinidiaceae; Actinidia
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 844aa MW: 92686.3 Da PI: 6.3466 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 58.4 | 1.2e-18 | 16 | 74 | 3 | 57 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHC....TS-HHHHHHHHHHHHHHHHC CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkkl....gLterqVkvWFqNrRakekk 57
k ++t+eq+e+Le+l++++++ps +r++L +++ +++ +q+kvWFqNrR +ek+
Achn049821 16 KYVRYTPEQVEALERLYHECPKPSSIRRQQLIRECpilsNIEPKQIKVWFQNRRCREKQ 74
5679*****************************************************97 PP
| |||||||
| 2 | START | 167.8 | 7.6e-53 | 160 | 374 | 2 | 205 |
HHHHHHHHHHHHHHHC-TT-EEEEEXCCTTEEEEEEESSS.SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-SEEEEEEEECTT....EEEEEEEE CS
START 2 laeeaaqelvkkalaeepgWvkssesengdevlqkfeeskvdsgealrasgvvdmvlallveellddkeqWdetlakaetlevissg....galqlmva 96
+aee+++e+++ka+ ++ Wv+++ +++g++++ +++ s++++g a+ra+g+v +++ v+e+l+d++ W ++++++++l+v+ ++ +l +m+
Achn049821 160 IAEETLAEFLSKATGTAVEWVQMPGMKPGPDSIGIVAISHGCTGVAARACGLVGLEPT-RVAEILKDRPSWFRDCRAVDVLNVLPTAnggtIELLYMQV 257
7899******************************************************.8888888888*****************9996633333333 PP
X....XTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--....-TTSEE-EESSEEEEEEEECTCEEEEEEEE-EE--SSXXHHHHHHHHHH CS
START 97 e....lqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppe...sssvvRaellpSgiliepksnghskvtwvehvdlkgrlphwllrslvks 187
l+a+++l+p Rdf+ +Ry+ +++g++v++++S++++q+ p +++vRae+lpSg+li+p+++g+s +++v+h++l++++++++lr+l++s
Achn049821 258 VlvfmLYAPTTLAPaRDFWLLRYTSVMEDGSLVVCERSLSNTQNGPAvppVQNFVRAEMLPSGYLIRPCEGGGSIIHIVDHMNLEAWSVPEVLRPLYES 356
222225789***********************************99988899*********************************************** PP
HHHHHHHHHHHHTXXXXX CS
START 188 glaegaktwvatlqrqce 205
+++ ++kt++a+l+++++
Achn049821 357 STVLAQKTTMAALRQLRQ 374
**************9876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50071 | 15.483 | 11 | 75 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.2E-15 | 13 | 79 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 5.56E-17 | 16 | 79 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 9.40E-17 | 16 | 76 | No hit | No description |
| Pfam | PF00046 | 3.0E-16 | 17 | 74 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 1.6E-18 | 18 | 74 | IPR009057 | Homeodomain-like |
| CDD | cd14686 | 1.15E-6 | 68 | 107 | No hit | No description |
| PROSITE profile | PS50848 | 25.437 | 150 | 371 | IPR002913 | START domain |
| CDD | cd08875 | 7.83E-77 | 154 | 376 | No hit | No description |
| SMART | SM00234 | 6.1E-39 | 159 | 375 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 5.49E-35 | 159 | 377 | No hit | No description |
| Gene3D | G3DSA:3.30.530.20 | 5.1E-22 | 159 | 372 | IPR023393 | START-like domain |
| Pfam | PF01852 | 2.4E-50 | 160 | 374 | IPR002913 | START domain |
| Pfam | PF08670 | 2.3E-52 | 700 | 842 | IPR013978 | MEKHLA |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0010014 | Biological Process | meristem initiation | ||||
| GO:0010075 | Biological Process | regulation of meristem growth | ||||
| GO:0010087 | Biological Process | phloem or xylem histogenesis | ||||
| GO:0048263 | Biological Process | determination of dorsal identity | ||||
| GO:0080060 | Biological Process | integument development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 844 aa Download sequence Send to blast |
MAMSCKDGKG VMDNGKYVRY TPEQVEALER LYHECPKPSS IRRQQLIREC PILSNIEPKQ 60 IKVWFQNRRC REKQRKEASR LQAVNRKLTA MNKLLMEEND RLQKQVSHLV YENGYFRQHT 120 QNTTLATKDT SCESVVTSGQ HHLTPQHPPR DASPAGLLSI AEETLAEFLS KATGTAVEWV 180 QMPGMKPGPD SIGIVAISHG CTGVAARACG LVGLEPTRVA EILKDRPSWF RDCRAVDVLN 240 VLPTANGGTI ELLYMQVVLV FMLYAPTTLA PARDFWLLRY TSVMEDGSLV VCERSLSNTQ 300 NGPAVPPVQN FVRAEMLPSG YLIRPCEGGG SIIHIVDHMN LEAWSVPEVL RPLYESSTVL 360 AQKTTMAALR QLRQIAQEVS QSNVTNWGRR PAALRALSQR LSRGFNEAVN GFTDEGWSMM 420 GNDGMDDVTI LVNSSPDKLM GLNLTFANGF PAVSNAVLCA KASMLLQNVP PAILLRFLRE 480 HRSEWADNNI DAYSAAAVKV GPCSLPGARV GSFGGQVILP LAHTIEHEEL LEVIKLEGVA 540 HCPEDAIMPR DMFLLQLCSG MDENAIGTCA ELIFAPIDAS FADDAPLLPS GFRIIPLDSG 600 KEASSPNRTL DLASALEIGP SGNKSSNDYT ANSGSTRSVM TIAFEFAFES HMQENVASMA 660 RQYVRSIISS VQRVALALSP SNLSSHGGLR TPLGTPEANT LARWICHSYR CFLGVELLTS 720 GSEGSETTLK TLWHHTDAIM CCSLKALPVF TFANQAGLDM LETTLVALQD ITLEKIFDDH 780 GRKTLCSEFP QIMQQGFACL QGGICLSSMG RPVSYERAVA WKVLNEEENA HCICFMFMNW 840 SFV* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of meristem development to promote lateral organ formation. May regulates procambial and vascular tissue formation or maintenance, and vascular development in inflorescence stems. {ECO:0000269|PubMed:15598805, ECO:0000269|PubMed:15705957, ECO:0000269|PubMed:16617092}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00200 | DAP | Transfer from AT1G52150 | Download |
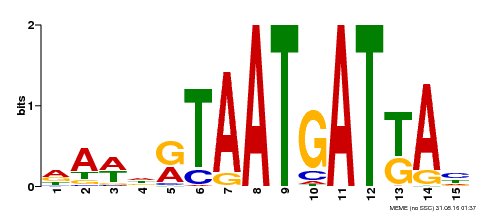
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By auxin. Repressed by miR165 and miR166. {ECO:0000269|PubMed:15773855, ECO:0000269|PubMed:16033795, ECO:0000269|PubMed:16617092, ECO:0000269|PubMed:17237362}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_028058078.1 | 0.0 | homeobox-leucine zipper protein ATHB-15 | ||||
| Swissprot | Q9ZU11 | 0.0 | ATB15_ARATH; Homeobox-leucine zipper protein ATHB-15 | ||||
| TrEMBL | A0A2R6R006 | 0.0 | A0A2R6R006_ACTCH; Homeobox-leucine zipper protein like | ||||
| STRING | VIT_09s0002g03740.t01 | 0.0 | (Vitis vinifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA457 | 24 | 140 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G52150.1 | 0.0 | HD-ZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



