 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Achn054811 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; Ericales; Actinidiaceae; Actinidia
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 721aa MW: 79289.9 Da PI: 6.6533 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 59.2 | 7e-19 | 26 | 81 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t++q++ Le+ F+ +++p++++r +L+++lgL rq+k+WFqNrR+++k
Achn054811 26 KKRYHRHTAYQIQKLEAIFKDSQHPDEKQRLQLSRELGLAPRQIKFWFQNRRTQMK 81
678899***********************************************998 PP
| |||||||
| 2 | START | 170.1 | 1.5e-53 | 222 | 448 | 2 | 206 |
HHHHHHHHHHHHHHHC-TT-EEEE....EXCCTTEEEEEEESSS.......SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S....EEEEEEEEC CS
START 2 laeeaaqelvkkalaeepgWvkss....esengdevlqkfeeskv......dsgealrasgvvdmvlallveellddkeqWdetla....kaetlevis 86
+a a++e++++ + +ep+W ks+ + +n d + + f+++++ ++ea+r+sgvv+m+ lv ++d++ +W e ++ k + lev+s
Achn054811 222 VAGNAMDEFISLLQNNEPLWIKSAndgrDVLNLDSYERLFPRANNhsknpnLRIEASRDSGVVIMNGLALVDMFMDSN-KWVELFPtivsKSQRLEVLS 319
67899*******************99998899999999999888899*******************************.******99999********* PP
TT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--..-TTSEE-EESSEEEEEEEECTCEEEEEEEE-EE--SSXX CS
START 87 sg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppe.sssvvRaellpSgiliepksnghskvtwvehvdlkgrlp 177
+g g lqlm+ elq+lsplvp R f+ +R+++q ++g+w+iv vS d +q +++ ss + ++++pSg+li++++ng+sk+twvehv+ ++++p
Achn054811 320 PGvlgnhsGSLQLMYEELQVLSPLVPtRQFYLLRFCQQIEQGSWAIVNVSYDLPQVNSQlSSLQALCHKFPSGCLIQDMPNGYSKITWVEHVEIEDKSP 418
**********************************************************99999999********************************* PP
.HHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 178 .hwllrslvksglaegaktwvatlqrqcek 206
h l+r l++sgla+ga w+ tlqr ce+
Achn054811 419 tHGLFRDLIHSGLAFGAELWLTTLQRMCER 448
****************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 4.8E-22 | 12 | 77 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 1.42E-19 | 13 | 83 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.718 | 23 | 83 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 3.3E-18 | 24 | 87 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 3.90E-18 | 25 | 84 | No hit | No description |
| Pfam | PF00046 | 2.1E-16 | 26 | 81 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 58 | 81 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 48.568 | 212 | 451 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 1.65E-33 | 213 | 449 | No hit | No description |
| CDD | cd08875 | 3.72E-113 | 216 | 447 | No hit | No description |
| SMART | SM00234 | 1.1E-45 | 221 | 448 | IPR002913 | START domain |
| Pfam | PF01852 | 2.4E-45 | 222 | 448 | IPR002913 | START domain |
| Gene3D | G3DSA:3.30.530.20 | 5.5E-5 | 227 | 257 | IPR023393 | START-like domain |
| Gene3D | G3DSA:3.30.530.20 | 5.5E-5 | 330 | 418 | IPR023393 | START-like domain |
| SuperFamily | SSF55961 | 1.33E-20 | 469 | 702 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009828 | Biological Process | plant-type cell wall loosening | ||||
| GO:0010091 | Biological Process | trichome branching | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 721 aa Download sequence Send to blast |
MEYGKGGGGR GSDGDDHDSA DPHRRKKRYH RHTAYQIQKL EAIFKDSQHP DEKQRLQLSR 60 ELGLAPRQIK FWFQNRRTQM KAQSERADNC ALWAENDRIR CENIAIREAL KNVICPTCGG 120 PPPVGEDSYF EEQKLRIENA HLKEELDRVS SMATKYIGRP ISQRLPVQPI HVSSLDLSMA 180 SFSGPPLDLD LLPGCSSTSV PNNPRFPAIN LSDMDKSLMA DVAGNAMDEF ISLLQNNEPL 240 WIKSANDGRD VLNLDSYERL FPRANNHSKN PNLRIEASRD SGVVIMNGLA LVDMFMDSNK 300 WVELFPTIVS KSQRLEVLSP GVLGNHSGSL QLMYEELQVL SPLVPTRQFY LLRFCQQIEQ 360 GSWAIVNVSY DLPQVNSQLS SLQALCHKFP SGCLIQDMPN GYSKITWVEH VEIEDKSPTH 420 GLFRDLIHSG LAFGAELWLT TLQRMCERFA CLMVSGNSTR DLGGVIPSPD GKRSIMKLAQ 480 RMVNNFCGSI NPSTGHQWTT LSGLNEVEVR SSLHRSTDPG QPSGVALSAA ATIWLPVSPQ 540 NVFNFFKDER TRPQWDVLFN GNAVQEVAHI ANGSHPGNCI SVLRAVNSNQ SNMLILQESC 600 IDSSGSLVVY CPVDLPAINI AMSGEDPSYI PLLPSGFTIL PDACHPHPHG GGPSTSSNPH 660 GGGGSLITVV FQILVSSYPS VKMNPESVGT VNNLIGTTVH QIKAALNCPP TTTTTITTSL 720 * |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor which acts as positive regulator of drought stress tolerance. Can transactivate CIPK3, NCED3 and ERECTA (PubMed:18451323). Transactivates several cell-wall-loosening protein genes by directly binding to HD motifs in their promoters. These target genes play important roles in coordinating cell-wall extensibility with root development and growth (PubMed:24821957). Transactivates CYP74A/AOS, AOC3, OPR3 and 4CLL5/OPCL1 genes by directly binding to HD motifs in their promoters. These target genes are involved in jasmonate (JA) biosynthesis, and JA signaling affects root architecture by activating auxin signaling, which promotes lateral root formation (PubMed:25752924). Acts as negative regulator of trichome branching (PubMed:16778018, PubMed:24824485). Required for the establishment of giant cell identity on the abaxial side of sepals (PubMed:23095885). May regulate cell differentiation and proliferation during root and shoot meristem development (PubMed:25564655). {ECO:0000269|PubMed:16778018, ECO:0000269|PubMed:18451323, ECO:0000269|PubMed:23095885, ECO:0000269|PubMed:24821957, ECO:0000269|PubMed:24824485, ECO:0000269|PubMed:25564655, ECO:0000269|PubMed:25752924}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00232 | DAP | Transfer from AT1G73360 | Download |
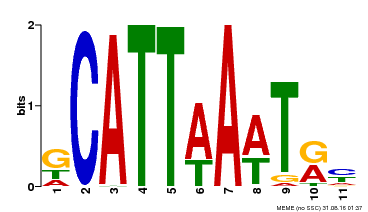
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_028078998.1 | 0.0 | homeobox-leucine zipper protein HDG11 | ||||
| Swissprot | Q9FX31 | 0.0 | HDG11_ARATH; Homeobox-leucine zipper protein HDG11 | ||||
| TrEMBL | A0A2R6PCJ1 | 0.0 | A0A2R6PCJ1_ACTCH; Homeobox-leucine zipper protein | ||||
| STRING | XP_010099679.1 | 0.0 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA2490 | 23 | 51 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73360.1 | 0.0 | homeodomain GLABROUS 11 | ||||



