 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Achn062591 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; Ericales; Actinidiaceae; Actinidia
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 304aa MW: 34057.6 Da PI: 7.6722 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 80.4 | 1.9e-25 | 129 | 190 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-ST...T---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsa...gCpvkkkversaedpkvveitYegeHnhe 59
++Dg++WrKYGqKe+ +++fpr Y+rCt++ gC ++k+v++ +e+p++ +itY g+H+++
Achn062591 129 MEDGHAWRKYGQKEILNAKFPRCYFRCTHKpdqGCLATKQVQKLQEEPHMYQITYFGHHTCT 190
58***************************9999***************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50811 | 18.337 | 124 | 187 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 4.3E-24 | 124 | 190 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 7.85E-24 | 126 | 190 | IPR003657 | WRKY domain |
| SMART | SM00774 | 6.3E-38 | 129 | 191 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 3.3E-24 | 130 | 189 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009862 | Biological Process | systemic acquired resistance, salicylic acid mediated signaling pathway | ||||
| GO:0009864 | Biological Process | induced systemic resistance, jasmonic acid mediated signaling pathway | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:1900056 | Biological Process | negative regulation of leaf senescence | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 304 aa Download sequence Send to blast |
MGTLRPEKLC ANRKRLMEEL VMGRDGAAQL RDLLEKPTGP GGSWPASAKG LVVKILGSFS 60 ASLSLLKCTS ETCEVCKIRA SVRVGSPSSG DRRSEDSGES SKRPAVKERR GCYKRRKTSD 120 TWMKILPTME DGHAWRKYGQ KEILNAKFPR CYFRCTHKPD QGCLATKQVQ KLQEEPHMYQ 180 ITYFGHHTCT NTLKISLECD PIKPYLLNFE SNNITSDQDH HHHPSYPISV KTECNKEETE 240 SNFSGKFPVV AAAAAAEPNM WADLLAMRSD HEDVISSTTT TSSHGLDMDF LNECVDFETD 300 FSW* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6ir8_A | 5e-22 | 125 | 189 | 4 | 68 | OsWRKY45 |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00408 | DAP | Transfer from AT3G56400 | Download |
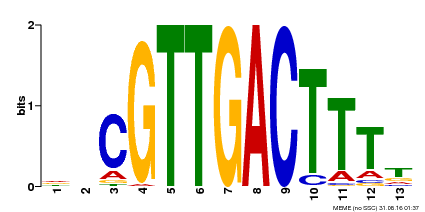
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_028079122.1 | 3e-97 | WRKY DNA-binding transcription factor 70-like | ||||
| TrEMBL | A0A2R6RTB3 | 0.0 | A0A2R6RTB3_ACTCH; WRKY transcription factor 70 | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA4230 | 21 | 43 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G56400.1 | 3e-48 | WRKY DNA-binding protein 70 | ||||



