 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Achn067431 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; Ericales; Actinidiaceae; Actinidia
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 243aa MW: 26381.5 Da PI: 7.4186 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 41.5 | 2.4e-13 | 190 | 235 | 6 | 55 |
HHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 6 nerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
+++Er RR ri +++ +L+el+P+ +k + a++Le+AveY+k Lq
Achn067431 190 SIAERIRRTRISDRIRKLQELVPNM----DKQTNTADMLEEAVEYVKFLQ 235
689*********************7....688*****************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50888 | 14.913 | 184 | 234 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 4.97E-15 | 187 | 239 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 6.89E-13 | 188 | 239 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 9.3E-15 | 189 | 239 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.2E-10 | 190 | 235 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 2.1E-12 | 190 | 240 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 243 aa Download sequence Send to blast |
MEPSAVGGGG EQSRGLARFR SAPATWLEAL LESEEEDDPL KPTQLLTQLP PTSSAASTNR 60 NSAAARYTDP GLFEGGRGAA SGLGFLRHNS SPADFLAQIS SGSDGYFSNF GIPSNYDDLS 120 SPMDVSPSSK RPKELDSERR KFSSQSKGEA GGQLQGGMSS LLDAEMEKLL EDSIPCRVRA 180 KRGCATHPRS IAERIRRTRI SDRIRKLQEL VPNMDKQTNT ADMLEEAVEY VKFLQKQIKI 240 KS* |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00181 | DAP | Transfer from AT1G35460 | Download |
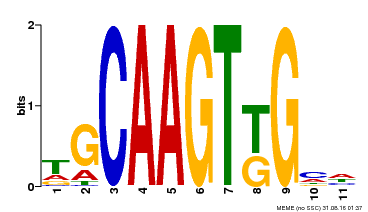
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_028059101.1 | 3e-98 | transcription factor bHLH80 isoform X1 | ||||
| Refseq | XP_028059102.1 | 3e-98 | transcription factor bHLH80 isoform X1 | ||||
| Swissprot | Q9C8P8 | 2e-79 | BH080_ARATH; Transcription factor bHLH80 | ||||
| TrEMBL | A0A2R6RPA2 | 1e-174 | A0A2R6RPA2_ACTCH; Transcription factor bHLH80 like | ||||
| STRING | EOY17321 | 4e-91 | (Theobroma cacao) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA11787 | 19 | 23 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G35460.1 | 3e-59 | bHLH family protein | ||||



