 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Achn079441 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; Ericales; Actinidiaceae; Actinidia
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 741aa MW: 80059.3 Da PI: 5.9882 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 100.8 | 8.4e-32 | 312 | 369 | 2 | 60 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS-- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhek 60
+D ynWrKYGqK+vkgse+prsYY+Ct+++Cpvkkkvers+ +++++ei+Y g Hnh+k
Achn079441 312 EDVYNWRKYGQKQVKGSEYPRSYYKCTHPNCPVKKKVERSH-EGHITEIIYDGAHNHPK 369
7****************************************.***************85 PP
| |||||||
| 2 | WRKY | 103.9 | 8.7e-33 | 528 | 586 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+vkg+++prsYY+CtsagC+v+k+ver+++d k v++tYeg+Hnh+
Achn079441 528 LDDGYRWRKYGQKVVKGNPNPRSYYKCTSAGCTVRKHVERASHDLKSVITTYEGKHNHD 586
59********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 4.0E-28 | 306 | 370 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 9.02E-25 | 309 | 370 | IPR003657 | WRKY domain |
| SMART | SM00774 | 5.6E-35 | 311 | 369 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 4.9E-24 | 312 | 368 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 23.146 | 312 | 370 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 5.5E-37 | 513 | 588 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.57E-28 | 520 | 588 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 38.48 | 523 | 588 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.2E-37 | 528 | 587 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 6.7E-25 | 529 | 586 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009555 | Biological Process | pollen development | ||||
| GO:0009942 | Biological Process | longitudinal axis specification | ||||
| GO:0030010 | Biological Process | establishment of cell polarity | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 741 aa Download sequence Send to blast |
MGGFDDHVAI IGDWVPPSPR SFFSALGDEI SSRSITELSI ENKNRSLFSG PQEKIISGRN 60 DENDGPRAGD QPAESNAFSE QKMNSRGGLL ERMAARAGFN APRLNTESIR PADLSLNPDV 120 RSPYLTIPPG LSPTTLLESP VFLSNSLVLP SPTTGKFPFS LSGNGKNSTL AEEAPGKSTE 180 NFFEDADTSS FAFKPVAASG PSLSLGPTGK MTPPNLPRQN FTGIEFSVQP ENSLKSQNVE 240 HSKVQYQDGT ALHLHQDFSR KDTAGNTIMS ESRAVGNGGG SGEHLSPHDD QQDEEADQRG 300 IGDTMVGGAS SEDVYNWRKY GQKQVKGSEY PRSYYKCTHP NCPVKKKVER SHEGHITEII 360 YDGAHNHPKP PPNRRSGSGP LYLSTDMQVD GADQPGAQIV AVGDPIWTTV QKGTAGGPLD 420 WRNENLEVTS SASVGAEYSN APNSLRGQNG THVEPGDEVD GSSTFSNEED EDDRATHGSI 480 SLGYDGEGDE SESKRRKIEA YATEMSGASR AIREPRVVVQ TTSEVDILDD GYRWRKYGQK 540 VVKGNPNPRS YYKCTSAGCT VRKHVERASH DLKSVITTYE GKHNHDIPAA RNSSHVNSGT 600 PNNGPAQSVG AAAVRTHLPR PEPSQFHNSM ARFDGHPVLS SLGFPGRQQL EPPPWFNFGM 660 NQTSLANLEM AGLGPNQGKL PVLPVYPYLG QQRLASEMGF VLPKGEPKVE PLSEPPLNLP 720 NGSSVYQQIT SRLPPLGPQM * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 2e-36 | 312 | 589 | 8 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 2e-36 | 312 | 589 | 8 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
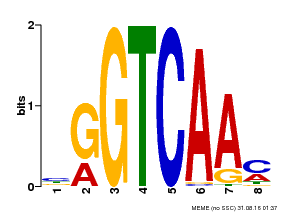
| UniProt | Transcription factor. Regulates WOX8 and WOX9 expression and basal cell division patterns during early embryogenesis. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element. Required to repolarize the zygote from a transient symmetric state. {ECO:0000269|PubMed:21316593}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00071 | PBM | Transfer from AT5G56270 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_028117015.1 | 0.0 | probable WRKY transcription factor 2 | ||||
| Swissprot | Q9FG77 | 0.0 | WRKY2_ARATH; Probable WRKY transcription factor 2 | ||||
| TrEMBL | A0A2R6P594 | 0.0 | A0A2R6P594_ACTCH; WRKY transcription factor | ||||
| STRING | XP_008246415.1 | 0.0 | (Prunus mume) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA3711 | 23 | 41 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G56270.1 | 0.0 | WRKY DNA-binding protein 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



