 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Achn095321 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; Ericales; Actinidiaceae; Actinidia
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 406aa MW: 45678.8 Da PI: 7.272 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 161.4 | 3.5e-50 | 21 | 175 | 2 | 128 |
NAM 2 ppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpk...........................kvkaeekewyfFskrdkkyatgkr 73
ppGfrFhPtdeel+++yL++kv + ++ i+evd++++ePwdLp +k +++ewyfFs rd+ky+tg r
Achn095321 21 PPGFRFHPTDEELITFYLAAKVFNGTFCG-VDIAEVDLNRCEPWDLPVrvrttlnqhpfefiiektnnsclccadVAKMGDREWYFFSMRDRKYPTGLR 118
8*************************877.34***************5589*************999999998875566999***************** PP
NAM 74 knratksgyWkatgkdkevlsk..kgelvglkktLvfykgrapkgektdWvmheyrl 128
+nrat +gyWkatgkd+ev+s+ +g l g+kktLvfykgrap+gekt+Wvmheyrl
Achn095321 119 TNRATGAGYWKATGKDREVYSSgpSGGLLGMKKTLVFYKGRAPRGEKTKWVMHEYRL 175
*********************9888889***************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 3.66E-55 | 13 | 65 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 55.246 | 20 | 197 | IPR003441 | NAC domain |
| Pfam | PF02365 | 2.0E-27 | 21 | 175 | IPR003441 | NAC domain |
| SuperFamily | SSF101941 | 3.66E-55 | 93 | 197 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010014 | Biological Process | meristem initiation | ||||
| GO:0010199 | Biological Process | organ boundary specification between lateral organs and the meristem | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 406 aa Download sequence Send to blast |
MLDVEDVLSE LNEDINEQGW PPGFRFHPTD EELITFYLAA KVFNGTFCGV DIAEVDLNRC 60 EPWDLPVRVR TTLNQHPFEF IIEKTNNSCL CCADVAKMGD REWYFFSMRD RKYPTGLRTN 120 RATGAGYWKA TGKDREVYSS GPSGGLLGMK KTLVFYKGRA PRGEKTKWVM HEYRLDGHFS 180 YRHMSKEEWV ICRIFHKKGE KKNPFYQGQS HLLEAAASPP TMGSLPPLLE AQSHNPMKAL 240 QNHPFPIHHH QDGDLKTLTN QVFPQSSFSP TPSPNNGINT NTNYHHPSPS MLFKSLASHQ 300 VCTLKEQPIT VPKQCKTEAN FYPFQLPDAN LRWLHRIHCD PHANPTLFLG MDFGGLMGFS 360 SATADATTTA AAPVSEMSTS IVFNNACVNP LIRSSVTGES WPLDL* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 3ulx_A | 2e-45 | 1 | 200 | 3 | 171 | Stress-induced transcription factor NAC1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator. Involved in molecular mechanisms regulating shoot apical meristem (SAM) formation during embryogenesis and organ separation. Required for axillary meristem initiation and separation of the meristem from the main stem. May act as an inhibitor of cell division. {ECO:0000269|PubMed:12837947, ECO:0000269|PubMed:17122068}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00241 | DAP | Transfer from AT1G76420 | Download |
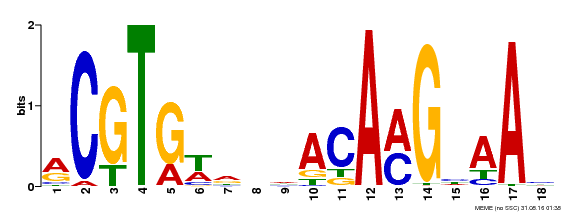
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By BRM, at the chromatin level, and conferring a very specific spatial expression pattern. {ECO:0000269|PubMed:16854978}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010663998.1 | 1e-153 | PREDICTED: protein CUP-SHAPED COTYLEDON 3 | ||||
| Swissprot | Q9S851 | 1e-88 | NAC31_ARATH; Protein CUP-SHAPED COTYLEDON 3 | ||||
| TrEMBL | A0A2R6PZA4 | 0.0 | A0A2R6PZA4_ACTCH; Protein CUP-SHAPED COTYLEDON 3 like | ||||
| STRING | EOY15912 | 1e-152 | (Theobroma cacao) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA152 | 24 | 279 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G76420.1 | 2e-90 | NAC family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



