 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Achn147111 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; Ericales; Actinidiaceae; Actinidia
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 424aa MW: 46994.1 Da PI: 9.695 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 33.8 | 7.8e-11 | 113 | 163 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHT.....TTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmg.....kgRtlkqcksrwqk 46
+++WT+eE+ l ++ ++G g W+tI + R+ ++k++w++
Achn147111 113 KQKWTPEEEAALRAGIVKHGAGKWRTILKDPEfssvlYLRSNVDLKDKWRN 163
79***********************************88***********9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 2.94E-15 | 110 | 179 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.6E-6 | 112 | 167 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.1E-13 | 113 | 164 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50090 | 7.48 | 113 | 165 | IPR017877 | Myb-like domain |
| Pfam | PF00249 | 1.3E-7 | 113 | 163 | IPR001005 | SANT/Myb domain |
| CDD | cd11660 | 8.32E-18 | 114 | 164 | No hit | No description |
| PROSITE profile | PS51504 | 23.326 | 222 | 296 | IPR005818 | Linker histone H1/H5, domain H15 |
| SuperFamily | SSF46785 | 5.28E-17 | 225 | 306 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SMART | SM00526 | 2.9E-12 | 226 | 291 | IPR005818 | Linker histone H1/H5, domain H15 |
| Gene3D | G3DSA:1.10.10.10 | 5.3E-15 | 227 | 298 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Pfam | PF00538 | 1.1E-7 | 233 | 289 | IPR005818 | Linker histone H1/H5, domain H15 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006334 | Biological Process | nucleosome assembly | ||||
| GO:0006357 | Biological Process | regulation of transcription from RNA polymerase II promoter | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0000786 | Cellular Component | nucleosome | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003691 | Molecular Function | double-stranded telomeric DNA binding | ||||
| GO:0033613 | Molecular Function | activating transcription factor binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| GO:0070491 | Molecular Function | repressing transcription factor binding | ||||
| GO:1990841 | Molecular Function | promoter-specific chromatin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 424 aa Download sequence Send to blast |
MVEGERGCRW WSLEEEGREV GKLDFLLDIE RKFLFTAHQP PPSLDTLTQT HNNNMSYYNQ 60 QPPVGVPPPQ GYPPEAYPKD AYPPPAYPPQ GTLGFVRLNL LRPFLPSLMG APKQKWTPEE 120 EAALRAGIVK HGAGKWRTIL KDPEFSSVLY LRSNVDLKDK WRNMSVMANG WGSREKARLA 180 LKRMQPISKQ DENSIALSPA GQSDEEIVDV KPLAVSTGSP QIGVPKRSMI RLDNLIMEAI 240 NGLKEPGGSN KTTIATYIEE EYWAPPNFKR LLSAKLKFLT ASGKLIKMKR KYRIAPPSEL 300 SGRRRKPSML LMEGRQKTSP KIDKDDINVL TKSQIDLELA KMRSMTPQEA AIAAAQAVAE 360 AEAAIAEAEE AAREAEAAEA DAEAAQAFAE AAMKTLKGRN TPSMSFSLSL SFAFRISLRF 420 CLI* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 288 | 305 | KRKYRIAPPSELSGRRRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds preferentially double-stranded telomeric repeats. {ECO:0000269|PubMed:19102728}. | |||||
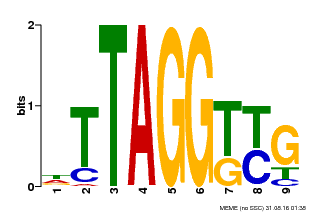
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00607 | ChIP-seq | Transfer from AT1G49950 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_028123872.1 | 1e-150 | telomere repeat-binding factor 1-like | ||||
| Swissprot | Q8VWK4 | 1e-103 | TRB1_ARATH; Telomere repeat-binding factor 1 | ||||
| TrEMBL | A0A2R6RGW7 | 0.0 | A0A2R6RGW7_ACTCH; Telomere repeat-binding factor like | ||||
| STRING | XP_006486912.1 | 1e-128 | (Citrus sinensis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA2864 | 24 | 48 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G49950.3 | 1e-103 | telomere repeat binding factor 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



