 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Achn182651 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; Ericales; Actinidiaceae; Actinidia
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 195aa MW: 21939.7 Da PI: 10.2113 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 99.1 | 1.8e-31 | 10 | 59 | 1 | 50 |
S---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE CS
SRF-TF 1 krienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklyeys 50
krien++nrqvtf+kRrng+lKKA+ELSvLCdaeva+i+fs++g+lyey+
Achn182651 10 KRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSTRGRLYEYA 59
79***********************************************8 PP
| |||||||
| 2 | K-box | 30.6 | 1.4e-11 | 78 | 130 | 4 | 69 |
K-box 4 ssgksleeakaeslqqelakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskK 69
s++ s++e +a+ +qqe+akL+ +i+nLq++ +eL++Le +Le+++++iRskK
Achn182651 78 SNTGSVSELNAQFYQQEAAKLRVQINNLQSS-------------NRELKNLEGRLERGISRIRSKK 130
3334499**********************98.............579******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50066 | 33.039 | 2 | 62 | IPR002100 | Transcription factor, MADS-box |
| SMART | SM00432 | 2.6E-40 | 2 | 61 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 6.17E-46 | 3 | 79 | No hit | No description |
| SuperFamily | SSF55455 | 2.22E-33 | 3 | 78 | IPR002100 | Transcription factor, MADS-box |
| PROSITE pattern | PS00350 | 0 | 4 | 58 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 4.8E-33 | 4 | 24 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 1.3E-26 | 11 | 58 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 4.8E-33 | 24 | 39 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 4.8E-33 | 39 | 60 | IPR002100 | Transcription factor, MADS-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0048440 | Biological Process | carpel development | ||||
| GO:0048443 | Biological Process | stamen development | ||||
| GO:0048497 | Biological Process | maintenance of floral organ identity | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 195 aa Download sequence Send to blast |
MGGRGKIEIK RIENTTNRQV TFCKRRNGLL KKAYELSVLC DAEVALIVFS TRGRLYEYAN 60 NSVKGTIERY KKASSGSSNT GSVSELNAQF YQQEAAKLRV QINNLQSSNR ELKNLEGRLE 120 RGISRIRSKK IAEGERVQQM NLMPGGSEYE LMQPPSFDTR NYLQVDGRLQ SDHNSYSRQD 180 QTVLQLVSSP TPNA* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1tqe_P | 9e-21 | 3 | 74 | 2 | 73 | Myocyte-specific enhancer factor 2B |
| 1tqe_Q | 9e-21 | 3 | 74 | 2 | 73 | Myocyte-specific enhancer factor 2B |
| 1tqe_R | 9e-21 | 3 | 74 | 2 | 73 | Myocyte-specific enhancer factor 2B |
| 1tqe_S | 9e-21 | 3 | 74 | 2 | 73 | Myocyte-specific enhancer factor 2B |
| 6c9l_A | 9e-21 | 3 | 74 | 2 | 73 | Myocyte-specific enhancer factor 2B |
| 6c9l_B | 9e-21 | 3 | 74 | 2 | 73 | Myocyte-specific enhancer factor 2B |
| 6c9l_C | 9e-21 | 3 | 74 | 2 | 73 | Myocyte-specific enhancer factor 2B |
| 6c9l_D | 9e-21 | 3 | 74 | 2 | 73 | Myocyte-specific enhancer factor 2B |
| 6c9l_E | 9e-21 | 3 | 74 | 2 | 73 | Myocyte-specific enhancer factor 2B |
| 6c9l_F | 9e-21 | 3 | 74 | 2 | 73 | Myocyte-specific enhancer factor 2B |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in regulating genes that determines stamen and carpel development in wild-type flowers. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00609 | ChIP-seq | Transfer from AT4G18960 | Download |
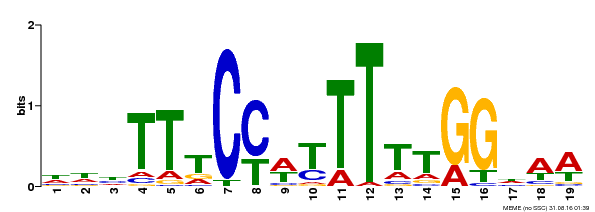
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HQ113361 | 1e-177 | HQ113361.1 Actinidia arguta AG mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_028062146.1 | 9e-98 | floral homeotic protein AGAMOUS-like | ||||
| Refseq | XP_028065683.1 | 1e-97 | floral homeotic protein AGAMOUS-like isoform X1 | ||||
| Refseq | XP_028065694.1 | 1e-97 | floral homeotic protein AGAMOUS-like isoform X1 | ||||
| Swissprot | Q40872 | 5e-90 | AG_PANGI; Floral homeotic protein AGAMOUS | ||||
| TrEMBL | F4ZKM4 | 1e-120 | F4ZKM4_ACTAR; AG | ||||
| STRING | EOY27872 | 8e-88 | (Theobroma cacao) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA40 | 24 | 625 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G18960.1 | 3e-81 | MIKC_MADS family protein | ||||



