 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Achn249361 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; Ericales; Actinidiaceae; Actinidia
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 492aa MW: 54530.5 Da PI: 6.8546 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 39.5 | 1.4e-12 | 163 | 219 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrd.pse.ng.krkrfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV++++++gr++A+++d ++ g rk + g ++ +e+Aa+a++ a++k++g
Achn249361 163 SQYRGVTRHRWTGRYEAHLWDnSCKkEGqTRKGRQ-GGYDMEEKAARAYDLAALKYWG 219
78*******************77776664346655.77******************98 PP
| |||||||
| 2 | AP2 | 35.8 | 2e-11 | 251 | 296 | 7 | 55 |
AP2 7 rwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
r++++ grW A+I + +k +lg+f t eeAa+a++ a+ k++g
Achn249361 251 RRHHQHGRWQARIGRVAG---NKDLYLGTFSTQEEAAEAYDIAAIKFRG 296
8999********988532...5************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 4.19E-17 | 163 | 227 | No hit | No description |
| SuperFamily | SSF54171 | 4.05E-13 | 163 | 228 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 8.0E-10 | 163 | 219 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 2.9E-20 | 164 | 233 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 4.7E-11 | 164 | 227 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 16.674 | 164 | 304 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 2.4E-6 | 165 | 176 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 2.10E-21 | 251 | 306 | No hit | No description |
| SMART | SM00380 | 1.8E-27 | 251 | 310 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 2.6E-6 | 251 | 296 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 1.05E-14 | 252 | 306 | IPR016177 | DNA-binding domain |
| Gene3D | G3DSA:3.30.730.10 | 2.4E-15 | 252 | 304 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 2.4E-6 | 286 | 306 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007276 | Biological Process | gamete generation | ||||
| GO:0010492 | Biological Process | maintenance of shoot apical meristem identity | ||||
| GO:0042127 | Biological Process | regulation of cell proliferation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 492 aa Download sequence Send to blast |
MAAHQYSSQE REAAMALTLD SLYYHQNPEI QQQEQEIEAQ NNPYYSGLPC NGMLYYSPLE 60 VQIKDNQMAT MGGGNWVSGH YSDHHGVVVD GSGGGGVSTC GDLQSLSLSM SPGSQSSCVI 120 SPSTGTDHCN NMAIETKKRG SAIVAPKQPV HRKSIDTFGQ RTSQYRGVTR HRWTGRYEAH 180 LWDNSCKKEG QTRKGRQGGY DMEEKAARAY DLAALKYWGP STHINCPLEN YQQELEEMKN 240 MTRQEYVAHL RRHHQHGRWQ ARIGRVAGNK DLYLGTFSTQ EEAAEAYDIA AIKFRGVNAV 300 TNFDISKYDV ERIMASNTLL SSELAKRNKD REPKSEAIEY HHHSSIQNNE ELVQSQSNTD 360 NNWKMVSCQS HSQSPQNPRF DSSLDEKLLC IGNYRNSSFL QDLIGIDSMS SGQAITMDEP 420 AKLGGVHFSN PSSLVTSLSS SREASPDKNG SMLFTKLPPT VGSWIPSAQL RAAAAPISVA 480 HLPVFAAWND T* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Acts as positive regulator of adventitious (crown) root formation by promoting its initiation. Promotes adventitious root initiation through repression of cytokinin signaling by positively regulating the two-component response regulator RR1. Regulated by the auxin response factor and transcriptional activator ARF23/ARF1. {ECO:0000269|PubMed:21481033}. | |||||
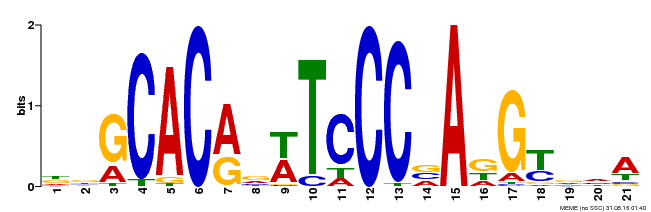
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00088 | SELEX | Transfer from AT4G37750 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by auxin. {ECO:0000269|PubMed:21481033}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002285467.2 | 0.0 | PREDICTED: AP2-like ethylene-responsive transcription factor ANT | ||||
| Swissprot | Q84Z02 | 1e-132 | CRL5_ORYSJ; AP2-like ethylene-responsive transcription factor CRL5 | ||||
| TrEMBL | A0A2R6RN93 | 0.0 | A0A2R6RN93_ACTCH; AP2-like ethylene-responsive transcription factor ANT | ||||
| STRING | VIT_18s0001g08610.t01 | 0.0 | (Vitis vinifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA2074 | 24 | 60 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G37750.1 | 1e-109 | AP2 family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



