 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Achn251771 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; Ericales; Actinidiaceae; Actinidia
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 382aa MW: 41867.3 Da PI: 6.9784 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 100.3 | 1.2e-31 | 219 | 274 | 1 | 56 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRla 56
k+r++W+peLH+rF++a++qLGGs++AtPk+i+elm v+gLt+++vkSHLQkYRl+
Achn251771 219 KARRCWSPELHRRFLHALQQLGGSHVATPKQIRELMMVDGLTNDEVKSHLQKYRLH 274
68****************************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 4.1E-28 | 216 | 277 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 13.953 | 216 | 276 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 4.18E-18 | 217 | 277 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 2.2E-26 | 219 | 274 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 9.6E-7 | 221 | 272 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 382 aa Download sequence Send to blast |
MINGYHNHNQ HNFTEKMQRC QEYIEALEKE RCKIQVFERE LPLCLDLVTQ AIESCRQQLS 60 GTTTTTEYFH GQSECSEQTS SEVPVLEQFI PIKRSSCSDD DEEQSEKPQN NNKDSKSNDK 120 SSKKSDWLKS VQLWNQTPDL PTKEDSPTNL PVIEVKKNGG GAFHPFKKEK NGGTIKTDPR 180 VLVPATAASS TTEIGGGDCG GSGDGDGKKE EKEGQSQRKA RRCWSPELHR RFLHALQQLG 240 GSHVATPKQI RELMMVDGLT NDEVKSHLQK YRLHTRRPTP STQNNNNPQQ PQFVVVGGIW 300 VPPPEYAAVA ASGEASGVAM TNGIYAPIAA PPPPPTQRQQ YNQSNQSHSG EMGSHSEGGG 360 GRCNSPATTS STHTSTASLA F* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4r_A | 3e-13 | 219 | 272 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 3e-13 | 219 | 272 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 3e-13 | 219 | 272 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 3e-13 | 219 | 272 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in phosphate signaling in roots. {ECO:0000250|UniProtKB:Q9FX67}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00160 | DAP | Transfer from AT1G25550 | Download |
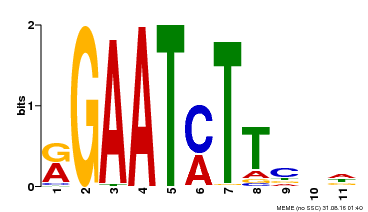
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003631224.1 | 1e-166 | PREDICTED: myb family transcription factor EFM | ||||
| Swissprot | Q9FPE8 | 1e-105 | HHO3_ARATH; Transcription factor HHO3 | ||||
| TrEMBL | A0A2R6PH37 | 0.0 | A0A2R6PH37_ACTCH; Myb family transcription factor EFM like | ||||
| STRING | VIT_01s0011g03110.t01 | 1e-163 | (Vitis vinifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA5219 | 24 | 38 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G25550.1 | 9e-83 | G2-like family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



