 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Achn274201 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; Ericales; Actinidiaceae; Actinidia
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 1065aa MW: 117927 Da PI: 8.8503 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 10.7 | 0.0017 | 977 | 995 | 5 | 23 |
TTTEEESSHHHHHHHHHHT CS
zf-C2H2 5 dCgksFsrksnLkrHirtH 23
Cgk F ++ +L++H r+H
Achn274201 977 GCGKKFFSHKYLVQHRRVH 995
7****************99 PP
| |||||||
| 2 | zf-C2H2 | 12.3 | 0.00052 | 1031 | 1057 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH..T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt..H 23
y+C Cg++F+ s++ rH r+ H
Achn274201 1031 YVCAeeGCGQTFRFVSDFSRHKRKtgH 1057
89********************99666 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00545 | 5.9E-13 | 26 | 67 | IPR003349 | JmjN domain |
| PROSITE profile | PS51183 | 13.786 | 27 | 68 | IPR003349 | JmjN domain |
| Pfam | PF02375 | 7.7E-14 | 28 | 61 | IPR003349 | JmjN domain |
| PROSITE profile | PS51184 | 32.57 | 196 | 365 | IPR003347 | JmjC domain |
| SMART | SM00558 | 1.0E-47 | 196 | 365 | IPR003347 | JmjC domain |
| SuperFamily | SSF51197 | 7.69E-26 | 210 | 370 | No hit | No description |
| Pfam | PF02373 | 5.0E-36 | 230 | 348 | IPR003347 | JmjC domain |
| SMART | SM00355 | 26 | 974 | 995 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 1.3E-4 | 978 | 1001 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 10.471 | 978 | 1000 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 1.14E-9 | 987 | 1029 | No hit | No description |
| PROSITE profile | PS50157 | 10.824 | 1001 | 1030 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0036 | 1001 | 1025 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 1.6E-8 | 1002 | 1025 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 1003 | 1025 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 3.24E-8 | 1019 | 1053 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 1.8E-9 | 1026 | 1054 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.85 | 1031 | 1057 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 11.032 | 1031 | 1062 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1033 | 1057 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009826 | Biological Process | unidimensional cell growth | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0033169 | Biological Process | histone H3-K9 demethylation | ||||
| GO:0035067 | Biological Process | negative regulation of histone acetylation | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1065 aa Download sequence Send to blast |
MASSSAPVAA EQPTTEVFPW LKTLPLAPEY HPTLAEFEDP IAYIFKIETE ASKYGICKIV 60 PPVAAPTKKI AIANLNRSLL ARSTTANPKS PPTFTTRQQQ IGFCPRKHRP VQKPVWQSGE 120 NYTFPQFEAK AKSFERNYLK KFPKKGLSAL EVETLYWKAF GDKPFSVEYA NDMPGSAFAP 180 VSARKKGRET GEAVTNVGET AWNMRGVSRA KGSLLRFMKE EIPGVTSPMA YLAMMFSWFA 240 WHVEDHDLHS LNYMHMGAGK TWYGVPREAA VAFEEVIRVH GYGGEINPLV TLAILGEKTT 300 VMSPEVLVGA GIPCCRLVQN AGEFVVTFPR AYHSGFSHGF NCGEAANIAT PGWLRVAKDA 360 AIRRAAINCP PMVSHFQLLY DLALSLCSRI PKSIDSEPRS SRLKHKRRGE GETLIKKLFA 420 EDVMKNNDLL HILAEGSSIV LLPHNSFDDS VYSNSRIASR FKGNSSFPLG LCNPEEEFKA 480 SEKFSSACDD QGLFSCVTCG ILSFACVAIV QPKDSAAHYL MSADCGIFND GDVVPRVTSD 540 GFTVAAGAAI LSRLDSRSGW VGKSTPDGLF DVPIQSADHI QTVDRSPELL SDTEIQKGTS 600 ALGLLALAYS SSDSEDEEVE ADIPLETHNI ASTESNSVGG RLTHQMKLST AASNDSPVAH 660 NALTSRSNTY VPMENTSMSF APKSGDDSYR PHVFCLQHAA EVEQQLRPIG GSHILLLCHP 720 DYPELEAEAK LVAEELAINY FWNDIAFREA TKEDEERIQT ALESEEGTHG NGDWAVKLGI 780 NLFYSANLGR SPLYSKQMPY NSIIYKAFGR TSLTNSPTKP KFHGKGSGRK KKIVVAGKWC 840 GKVWMSNQVH PLLVERDAEE GEDDNTSFRG WAKPEVKRER LSERARLAET NFEIKKIGKK 900 RKNTTQKEGG AIKKSKSAKV EDSSPQTQRS SDCNKSRFDS FIEDELEGGP STRLRKRTSK 960 LPAEDKLVLK KHTSIKGCGK KFFSHKYLVQ HRRVHMDDRP LKCPWKGCKM AFKWAWARTE 1020 HIRVHTGARP YVCAEEGCGQ TFRFVSDFSR HKRKTGHSTK KGRG* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6ip0_A | 3e-74 | 15 | 386 | 3 | 353 | Transcription factor jumonji (Jmj) family protein |
| 6ip4_A | 3e-74 | 15 | 386 | 3 | 353 | Arabidopsis JMJ13 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Histone demethylase that demethylates 'Lys-27' (H3K27me) of histone H3. Demethylates both tri- (H3K27me3) and di-methylated (H3K27me2) H3K27me (PubMed:21642989, PubMed:27111035). Demethylates also H3K4me3/2 and H3K36me3/2 in an in vitro assay (PubMed:20711170). Involved in the transcriptional regulation of hundreds of genes regulating developmental patterning and responses to various stimuli (PubMed:18467490). Binds DNA via its four zinc fingers in a sequence-specific manner, 5'-CTCTGYTY-3', to promote the demethylation of H3K27me3 and the regulation of organ boundary formation (PubMed:27111034, PubMed:27111035). Involved in the regulation of flowering time by repressing FLOWERING LOCUS C (FLC) expression (PubMed:15377760). Interacts with the NF-Y complexe to regulate SOC1 (PubMed:25105952). Mediates the recruitment of BRM to its target loci (PubMed:27111034). {ECO:0000269|PubMed:15377760, ECO:0000269|PubMed:18467490, ECO:0000269|PubMed:20711170, ECO:0000269|PubMed:21642989, ECO:0000269|PubMed:25105952, ECO:0000269|PubMed:27111034, ECO:0000269|PubMed:27111035}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00608 | ChIP-seq | Transfer from AT3G48430 | Download |
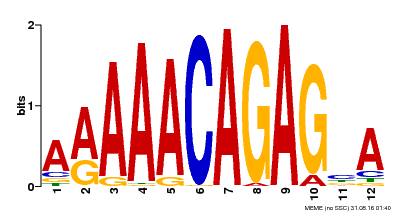
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009613169.1 | 0.0 | PREDICTED: lysine-specific demethylase REF6 isoform X2 | ||||
| Refseq | XP_016435657.1 | 0.0 | PREDICTED: lysine-specific demethylase REF6-like isoform X2 | ||||
| Swissprot | Q9STM3 | 0.0 | REF6_ARATH; Lysine-specific demethylase REF6 | ||||
| TrEMBL | F6GTM2 | 0.0 | F6GTM2_VITVI; Uncharacterized protein | ||||
| STRING | VIT_17s0000g09980.t01 | 0.0 | (Vitis vinifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA4988 | 23 | 31 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48430.1 | 0.0 | relative of early flowering 6 | ||||



