 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Achn314301 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; Ericales; Actinidiaceae; Actinidia
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 689aa MW: 76020 Da PI: 7.8751 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 105.7 | 2.4e-33 | 344 | 401 | 2 | 60 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS-- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhek 60
+DgynWrKYGqK+vkgse+prsYY+Ct+++Cp+kkkvers d++++ei+Y+g+Hnh+k
Achn314301 344 EDGYNWRKYGQKQVKGSENPRSYYKCTFQNCPTKKKVERSL-DGQITEIVYKGNHNHPK 401
7****************************************.***************85 PP
| |||||||
| 2 | WRKY | 105.2 | 3.5e-33 | 512 | 570 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+vkg+++prsYY+Ct +gCpv+k+ver+++d ++v++tYeg+Hnh+
Achn314301 512 LDDGYRWRKYGQKVVKGNPNPRSYYKCTNPGCPVRKHVERASHDLRAVITTYEGKHNHD 570
59********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 1.1E-28 | 333 | 402 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 23.973 | 338 | 402 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 2.22E-25 | 339 | 402 | IPR003657 | WRKY domain |
| SMART | SM00774 | 8.8E-37 | 343 | 401 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 2.3E-25 | 344 | 400 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 2.5E-37 | 497 | 572 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 3.92E-29 | 504 | 572 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 38.377 | 507 | 572 | IPR003657 | WRKY domain |
| SMART | SM00774 | 3.1E-39 | 512 | 571 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.2E-25 | 513 | 570 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0009788 | Biological Process | negative regulation of abscisic acid-activated signaling pathway | ||||
| GO:0009938 | Biological Process | negative regulation of gibberellic acid mediated signaling pathway | ||||
| GO:0010120 | Biological Process | camalexin biosynthetic process | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0010508 | Biological Process | positive regulation of autophagy | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0070370 | Biological Process | cellular heat acclimation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 689 aa Download sequence Send to blast |
MCMHDNLDSV VQIAPKMTFS VSAGTFLTSR FNPLYAFLGF YESFINLSYF SVIDSGLLAP 60 ASISGCFDSK WSCSCPVGTW STYRSEAPDI VNVLHLSLIT LKLLVVCSSR GIKHFSLDTS 120 GNSHPQPPPP SHFSFMSSSA NNTSFSDLLA GDYPSAAPVS RGLSDRIAER SGSGVPKFKS 180 ITPPSLPISP PPISPFSYFA IPPGLSPAEL LDSPVLLSTS NVLPSPTTGS FPTHAFNWKS 240 NSNHYHNQRS VKQEQKNFSD FSFHSQKGPP TSSATSFQSQ VPAENKTWGF QETTKQVKPE 300 SASMQSFSPE ISTIQNNSQS GFQSDYNNHY NQSSQSIREQ RRSEDGYNWR KYGQKQVKGS 360 ENPRSYYKCT FQNCPTKKKV ERSLDGQITE IVYKGNHNHP KPQSTRRSAA AASSQAIQPH 420 TNEIPDQSYA SHGNGPMDSV ATPEISSISM GDDDYDQNSQ KSKSGGDEFD EEEPDAKRWK 480 IEGESEGISA PGSRTVREPR VVVQTTSDID ILDDGYRWRK YGQKVVKGNP NPRSYYKCTN 540 PGCPVRKHVE RASHDLRAVI TTYEGKHNHD VPAARGSGSH SVNRPLPTAT TTNHHHHQQA 600 TAIRPSPMFH HSSTHAMANP IRNSRLQAPD QGQAPFTLEM LQSPGSFGFS GFENSMGSYM 660 NQNQLADNVF SRTKEEPRDD LFVESLLY* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 5e-37 | 335 | 573 | 8 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 5e-37 | 335 | 573 | 8 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription repressor (By similarity). Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element. Negative regulator of both gibberellic acid (GA) and abscisic acid (ABA) signaling in aleurone cells, probably by interfering with GAM1, via the specific repression of GA- and ABA-induced promoters (By similarity). {ECO:0000250|UniProtKB:Q6IEQ7, ECO:0000250|UniProtKB:Q6QHD1}. | |||||
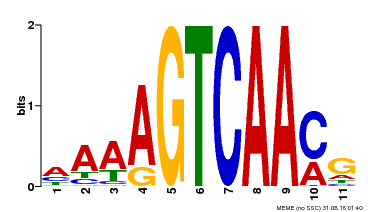
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00299 | DAP | Transfer from AT2G38470 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by abscisic acid (ABA) in aleurone cells, embryos, roots and leaves (PubMed:25110688). Slightly down-regulated by gibberellic acid (GA) (By similarity). Accumulates in response to jasmonic acid (MeJA) (PubMed:16919842). {ECO:0000250|UniProtKB:Q6IEQ7, ECO:0000269|PubMed:16919842, ECO:0000269|PubMed:25110688}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_028051918.1 | 0.0 | WRKY transcription factor WRKY24-like | ||||
| Swissprot | Q6B6R4 | 1e-134 | WRK24_ORYSI; WRKY transcription factor WRKY24 | ||||
| TrEMBL | A0A2R6QJE6 | 0.0 | A0A2R6QJE6_ACTCH; WRKY transcription factor | ||||
| STRING | EOY34631 | 0.0 | (Theobroma cacao) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA2286 | 24 | 60 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G38470.1 | 1e-106 | WRKY DNA-binding protein 33 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



