 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aco000069.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Bromeliaceae; Ananas
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 622aa MW: 66405.6 Da PI: 6.473 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 102.6 | 2.3e-32 | 263 | 320 | 2 | 60 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS-- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhek 60
+DgynWrKYGqK+vkgse+prsYY+Ct+++C+v+k++ers+ d++++ei+Y+g+H+h+k
Aco000069.1 263 EDGYNWRKYGQKHVKGSENPRSYYKCTHPNCNVRKQLERSH-DGQITEIIYKGKHDHPK 320
8****************************************.***************85 PP
| |||||||
| 2 | WRKY | 105.9 | 2.2e-33 | 435 | 493 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+vkg+++prsYY+Ct gCpv+k+ver+++dpk+v++tYeg+Hnh+
Aco000069.1 435 LDDGYRWRKYGQKVVKGNPNPRSYYKCTNIGCPVRKHVERASHDPKAVITTYEGKHNHD 493
59********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 2.8E-28 | 260 | 321 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 7.32E-25 | 261 | 321 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.7E-35 | 262 | 320 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 24.501 | 263 | 321 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 2.0E-24 | 263 | 319 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 1.5E-37 | 420 | 495 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.14E-29 | 427 | 495 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 38.733 | 430 | 495 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.5E-39 | 435 | 494 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 3.9E-26 | 436 | 493 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009961 | Biological Process | response to 1-aminocyclopropane-1-carboxylic acid | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 622 aa Download sequence Send to blast |
MEESPANAGD LACGGREGAT PAPELGSEGL AAGGGGSGSV GEGAAVHPRV SSLAAASAAR 60 YKAMSPGRLP IATRAPCLTI PQGFSPSALL ESPVLLTNIK ARDLTPFSLP SKAEPSPTTG 120 TLNISYMMNR SVHSSKLSSP RETSNSVYSD KSSGDFEFKL HAGSSLHSAV PSLLPQDSAD 180 GKLEECEASV QIQSQVQNHK SAPVIPQTAL TQPIENITLK AGVHSEIACD ELQQIDSSDK 240 TMQELQSNLS GSAPLPVLEK SAEDGYNWRK YGQKHVKGSE NPRSYYKCTH PNCNVRKQLE 300 RSHDGQITEI IYKGKHDHPK PQPNRRSAVG VLSSQGEEKA DSLVNAEDKS SNALDHLCNH 360 VNPIVTPELS PASASEDEVD VGCGQSNTID EVADDDDPES KRRKMDITTI DAATIGKPNR 420 EPRVVVQTVS EVDILDDGYR WRKYGQKVVK GNPNPRSYYK CTNIGCPVRK HVERASHDPK 480 AVITTYEGKH NHDVPAARNA SHEISAPIPM VTNSSNLISN RMPAALSSIL SIGNPRVTSH 540 QYTQLAGSDT VSLDLGVGIN PNHGNAGNEI QSLGSNQIQH HQSTLYGSRE DDGEGFTFKA 600 TPMNHSANMY YTSAGNLVLG P* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 1e-37 | 263 | 496 | 8 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 1e-37 | 263 | 496 | 8 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in starch synthesis (PubMed:12953112). Acts as a transcriptional activator in sugar signaling (PubMed:16167901). Interacts specifically with the SURE and W-box elements, but not with the SP8a element (PubMed:12953112). {ECO:0000269|PubMed:12953112, ECO:0000269|PubMed:16167901}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00455 | DAP | Transfer from AT4G26640 | Download |
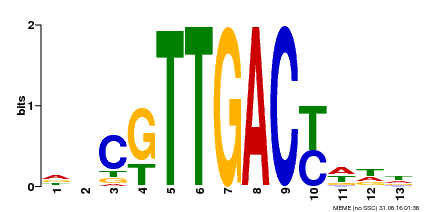
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by sugar. {ECO:0000269|PubMed:12953112}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020100227.1 | 0.0 | WRKY transcription factor SUSIBA2-like | ||||
| Swissprot | Q6VWJ6 | 0.0 | WRK46_HORVU; WRKY transcription factor SUSIBA2 | ||||
| TrEMBL | A0A199VQY2 | 0.0 | A0A199VQY2_ANACO; WRKY transcription factor SUSIBA2 | ||||
| STRING | XP_008800638.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6740 | 38 | 51 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G26640.2 | 1e-127 | WRKY family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aco000069.1 |



