 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aco000773.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Bromeliaceae; Ananas
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 459aa MW: 50278.8 Da PI: 9.2846 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 39.2 | 1.2e-12 | 385 | 430 | 6 | 55 |
HHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 6 nerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
+++Er RR ri +++ +L+el+P+ +k + a++L +Av+YIk+Lq
Aco000773.1 385 SIAERVRRTRISERMRKLQELVPNM----DKQTNTADMLDLAVDYIKDLQ 430
689*********************7....688*****************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50888 | 15.25 | 379 | 429 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 3.14E-16 | 382 | 441 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 6.09E-13 | 383 | 434 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 4.1E-15 | 384 | 441 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 9.5E-14 | 385 | 435 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.3E-9 | 385 | 430 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0048573 | Biological Process | photoperiodism, flowering | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 459 aa Download sequence Send to blast |
MYGSPQAARS KDLNLPVSAE PAFGHRKEEK ELLQRHHHHH HHRDHQQHQH HNPPPPPPPP 60 HHQQQQQQQQ QMSSGLLRYR SAPTALLGEV VDDFLPVRPS SPETETMFAR FLATSDLRDE 120 IREKPPSAAA QRSSTFASAM DEAAASQQQQ QQQQQQMMYH SHHQPMPSHG SVESLYRTAS 180 SAAIDLEPMK SDGATNNLIR QSSSPPGIFS HVNVDNGYSV MRGIGGFRNG NNSLGAVNAA 240 NTNSTSNRMK GQISFSSRQG SLMSQISGMG SEGGAIGGSS PEDGGNSGGT AAGGGRYIPG 300 YPLGSWDDPP LLSDHFSGSL KRGREAEGKI ISGLNQSEHQ NGLMHQFSLP KTSSEMAAIE 360 KFLQFHDAVP CKIRAKRGCA THPRSIAERV RRTRISERMR KLQELVPNMD KQTNTADMLD 420 LAVDYIKDLQ KQVKTLTENR ASCTCSSSKQ KQYQNPAA* |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00646 | PBM | Transfer from LOC_Os08g39630 | Download |
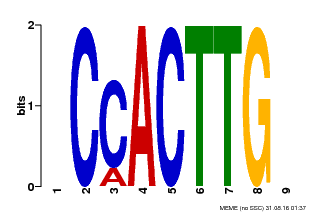
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KM662043 | 0.0 | KM662043.1 Ananas bracteatus clone 45301b microsatellite sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020082673.1 | 0.0 | transcription factor bHLH130-like | ||||
| TrEMBL | A0A199VBP5 | 0.0 | A0A199VBP5_ANACO; Transcription factor bHLH130 | ||||
| STRING | XP_008776552.1 | 1e-168 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1884 | 38 | 103 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G42280.1 | 3e-70 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aco000773.1 |



