 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aco000913.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Bromeliaceae; Ananas
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 488aa MW: 53177.2 Da PI: 6.3323 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 27.4 | 6.2e-09 | 312 | 360 | 3 | 55 |
HHHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 3 rahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
+h+ +Er RR++i +++ L++l+P + sk Ka +L + ++Y++sLq
Aco000913.1 312 NSHSLAERLRREKISERMKFLQDLVPGC----SKVTGKAVMLDEIINYVQSLQ 360
58*************************9....556*****************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 5.32E-11 | 307 | 364 | No hit | No description |
| SuperFamily | SSF47459 | 6.41E-17 | 307 | 378 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 15.51 | 309 | 359 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 1.8E-16 | 310 | 376 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 3.0E-6 | 312 | 360 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 8.6E-10 | 315 | 365 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 488 aa Download sequence Send to blast |
MDMNEKEKLG LEKNEDHMSY HASDISTAWQ QFNSSSGAIG QEIQMGSATC PSNPMVDSFN 60 PSLWNHHPSS QNLVGLSENN AHVGNPPMGL NPSSFFPKGG TFLPSLPKIP PPPILPHFSA 120 DTGFIERAAR FSCFGSGNLS GMVNPFGASE ILSPYANPSK FSGQKSEMSV DEASENAPLP 180 VNDHLSSIRS PRERRDDANS LRESAEPEFS GGGEEEAPNL ATAARDNSSS KGPNAKKRKR 240 SNQGGAETEQ VEGAPQVPIE TTKENMDNKQ KNEQNSSTVG TSKASGKQAK DNSDLPKEDY 300 IHVRARRGQA TNSHSLAERL RREKISERMK FLQDLVPGCS KVTGKAVMLD EIINYVQSLQ 360 RQVEFLSMKL AAVNPRLDFN IEGLLSKELF SRGGPSSAIG FSPDMIHPHL HPSQHGMVQA 420 GMPSIANPSD VLRRVMNAQP TSINGYKEPT SQMPNTWDEE LQNVMQMSYP PLNAQELNSK 480 PRDGFPL* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 235 | 240 | KKRKRS |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00660 | PBM | Transfer from Pp3c6_9520 | Download |
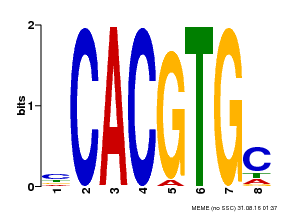
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020114802.1 | 0.0 | transcription factor bHLH49 | ||||
| TrEMBL | A0A199VA48 | 0.0 | A0A199VA48_ANACO; Transcription factor bHLH49 | ||||
| STRING | XP_008788970.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2909 | 37 | 85 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G68920.2 | 3e-90 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aco000913.1 |



