 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aco003842.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Bromeliaceae; Ananas
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 586aa MW: 62563.5 Da PI: 9.2389 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 105.9 | 2.1e-33 | 303 | 358 | 2 | 58 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnh 58
dDgynWrKYGqK+vkgse+prsYY+Ct ++Cpvkkkvers d++++ei+Y+g+Hnh
Aco003842.1 303 DDGYNWRKYGQKMVKGSEYPRSYYKCTNPNCPVKKKVERSV-DGQITEIIYKGQHNH 358
8****************************************.*************** PP
| |||||||
| 2 | WRKY | 105.7 | 2.3e-33 | 468 | 526 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+vkg+++prsYY+Ct+agC+v+k++er+++dp++v++tYeg+Hnh+
Aco003842.1 468 LDDGYRWRKYGQKVVKGNPHPRSYYKCTYAGCNVRKHIERASTDPRAVITTYEGKHNHD 526
59********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 1.3E-29 | 297 | 360 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 7.98E-26 | 300 | 361 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.2E-37 | 302 | 360 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.5E-25 | 303 | 358 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 24.122 | 303 | 361 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 6.5E-37 | 454 | 528 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 2.09E-29 | 460 | 528 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 38.251 | 463 | 528 | IPR003657 | WRKY domain |
| SMART | SM00774 | 4.5E-39 | 468 | 527 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 5.5E-26 | 469 | 526 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 586 aa Download sequence Send to blast |
MTILPLAIVV FFPIFLSRPF IRSRFPSQSA AISLHFLLHL LPFSSCTLAA LDFGDSRRNR 60 EGARRRPPRG RRSRCRARAA YESLFAGGGG GGGGGGGGVA AAAAEVSPGP LTLASSLFPD 120 ESESECRSFT QLLIGAMSSP VGGGGGGGGL ERRGGGGGGG GGGGGGGGLV RFGQNRPTSL 180 AVTQPPVFMI PPGLSPATLL DSPAFLSPMG NFGMSHQQAL AQVTAQAAHS QFKIHSQPEY 240 PSLLSIPATT CTIHHNTSAV NMAPIPDIST LSTNSNNTKL ESAEASQADQ RFQPAHVVDK 300 PADDGYNWRK YGQKMVKGSE YPRSYYKCTN PNCPVKKKVE RSVDGQITEI IYKGQHNHLR 360 PPNRRSKEGG ALQSESEEFN ENAEKTGISN DNLLASSIGK RDREPSYGTH EQLSGSSDSE 420 EVGGGEVKAD EADNEEPDAK RRNIATSSQR ALTEPRIIVQ TTSEVDLLDD GYRWRKYGQK 480 VVKGNPHPRS YYKCTYAGCN VRKHIERAST DPRAVITTYE GKHNHDVPAA RSSSHSIASA 540 SVSSSGLQNR GQNTHNQASL NRTDFGSRDH RAAAVLQLKE ENDIT* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 9e-40 | 303 | 529 | 8 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 9e-40 | 303 | 529 | 8 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (By similarity). {ECO:0000250}. | |||||
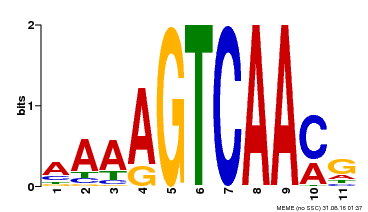
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00260 | DAP | Transfer from AT2G03340 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salicylic acid and during leaf senescence. {ECO:0000269|PubMed:11449049, ECO:0000269|PubMed:11722756}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KM662594 | 1e-177 | KM662594.1 Ananas bracteatus clone 46433 microsatellite sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020104924.1 | 0.0 | probable WRKY transcription factor 3 | ||||
| Swissprot | Q9ZQ70 | 1e-140 | WRKY3_ARATH; Probable WRKY transcription factor 3 | ||||
| TrEMBL | A0A199USJ9 | 0.0 | A0A199USJ9_ANACO; Putative WRKY transcription factor 3 | ||||
| STRING | XP_008790093.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1345 | 38 | 122 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G13960.1 | 1e-122 | WRKY DNA-binding protein 4 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aco003842.1 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



