 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aco004208.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Bromeliaceae; Ananas
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 606aa MW: 66745.5 Da PI: 10.9058 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 63.6 | 4e-20 | 437 | 486 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrd.psengkrkrfslgkfgtaeeAakaaiaarkkleg 55
+++GVr ++ +gr++AeIrd ++ ++r +lg+f+taeeAa+a+++a+++l+g
Aco004208.1 437 HFRGVRKRP-WGRFAAEIRDpWK----KTRKWLGTFDTAEEAARAYDEAARNLRG 486
69*******.**********883....5*************************99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF54171 | 1.05E-20 | 437 | 494 | IPR016177 | DNA-binding domain |
| CDD | cd00018 | 1.25E-26 | 437 | 493 | No hit | No description |
| SMART | SM00380 | 2.6E-34 | 437 | 500 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 8.5E-30 | 437 | 493 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 22.273 | 437 | 494 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.6E-9 | 438 | 449 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.6E-9 | 460 | 476 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 606 aa Download sequence Send to blast |
MSCSRPVRVL EPSYRFRWRF NVTPNYSLTP PPLSTTTHPV RGQFGHFGTP ETPRLGDDDD 60 ADADAICVEI CASDASAALI RAHHDTKQPG PSGLNPRGGE GGGFYRKVLS LLINSSDRST 120 ARFGAVTKTD GGSSAIQCCS SSMMMVAHGT QKQQKQQHHY QGVVIRGIMR ELSPDRILKP 180 VGPTPPNPPL VTFTARGAGK VRSPNEDKRE KDVGVVADRR PPHLNPTRRP QAAWTHQIRR 240 RRNGMCVWEG DIRHDKGSRE GDRTTLLHTR ERPSRVHHYR RMHKDSGKRW RWWWWWWWFV 300 GRGVGTKTKR KGKEKRKEKG ERRAWAGAGF SGEVVATRAA AAAAAVTASP AAVPSTLSFD 360 LFPPGPTLLH TVLSAAPLGL SRTGGEECVA RGEEREQEQE QRQRQGQGQG QGHLSLAMAV 420 EMLRLGKDGF ASGKEVHFRG VRKRPWGRFA AEIRDPWKKT RKWLGTFDTA EEAARAYDEA 480 ARNLRGPKAK TNFGYVGSDA DIPSAIPVRG GAVSVAASAG APQVGWWRSA FAAAAPDGRD 540 LILGPPGASG GSDFSGYRFD AVEVVVRGEQ QKKKMMAAMA RKEEEANERR KKPFSFDLNL 600 PAPLF* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gcc_A | 3e-17 | 437 | 493 | 2 | 59 | ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 |
| 2gcc_A | 3e-17 | 437 | 493 | 5 | 62 | ATERF1 |
| 3gcc_A | 3e-17 | 437 | 493 | 5 | 62 | ATERF1 |
| 5wx9_A | 1e-16 | 438 | 493 | 15 | 71 | Ethylene-responsive transcription factor ERF096 |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 309 | 317 | RKGKEKRKE |
| 2 | 571 | 591 | KKKMMAAMARKEEEANERRKK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00194 | DAP | Transfer from AT1G50640 | Download |
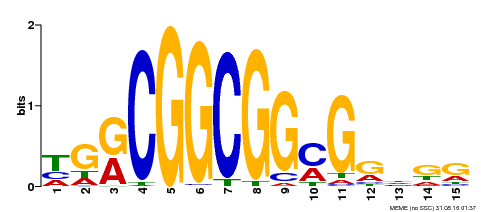
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP103 | 37 | 465 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G50640.1 | 8e-22 | ethylene responsive element binding factor 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aco004208.1 |



